Dysregulated Provision of Oxidisable Substrates to the Mitochondria in ME/CFS Lymphoblasts - PubMed (original) (raw)
Dysregulated Provision of Oxidisable Substrates to the Mitochondria in ME/CFS Lymphoblasts
Daniel Missailidis et al. Int J Mol Sci. 2021.
Abstract
Although understanding of the biomedical basis of Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) is growing, the underlying pathological mechanisms remain uncertain. We recently reported a reduction in the proportion of basal oxygen consumption due to ATP synthesis by Complex V in ME/CFS patient-derived lymphoblast cell lines, suggesting mitochondrial respiratory inefficiency. This was accompanied by elevated respiratory capacity, elevated mammalian target of rapamycin complex 1 (mTORC1) signaling activity and elevated expression of enzymes involved in the TCA cycle, fatty acid β-oxidation and mitochondrial transport. These and other observations led us to hypothesise the dysregulation of pathways providing the mitochondria with oxidisable substrates. In our current study, we aimed to revisit this hypothesis by applying a combination of whole-cell transcriptomics, proteomics and energy stress signaling activity measures using subsets of up to 34 ME/CFS and 31 healthy control lymphoblast cell lines from our growing library. While levels of glycolytic enzymes were unchanged in accordance with our previous observations of unaltered glycolytic rates, the whole-cell proteomes of ME/CFS lymphoblasts contained elevated levels of enzymes involved in the TCA cycle (p = 1.03 × 10-4), the pentose phosphate pathway (p = 0.034, G6PD p = 5.5 × 10-4), mitochondrial fatty acid β-oxidation (p = 9.2 × 10-3), and degradation of amino acids including glutamine/glutamate (GLS p = 0.034, GLUD1 p = 0.048, GOT2 p = 0.026), branched-chain amino acids (BCKDHA p = 0.028, BCKDHB p = 0.031) and essential amino acids (FAH p = 0.036, GCDH p = 0.006). The activity of the major cellular energy stress sensor, AMPK, was elevated but the increase did not reach statistical significance. The results suggest that ME/CFS metabolism is dysregulated such that alternatives to glycolysis are more heavily utilised than in controls to provide the mitochondria with oxidisable substrates.
Keywords: ME/CFS; Myalgic Encephalomyelitis; TCA cycle; amino acid catabolism; beta-oxidation; glycolysis; metabolism; mitochondria; proteomics; transcriptomics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
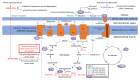
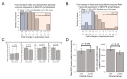
Figure 1
Simplified depiction of oxidisable substrate provision and usage by the mitochondria. The generalised “flow” of substrate molecules derived from glucose, fatty acids or amino acids is represented by arrows colour-coded green, blue and red, respectively. Reducing equivalents are denoted by purple. Processes are italicised. Glucose may be catabolised by glycolysis in order to provision the mitochondria with pyruvate, converted by pyruvate dehydrogenase (PDH) to acetyl CoA for entry into the TCA cycle. Fatty acid β-oxidation also provides acetyl CoA for the TCA cycle, by the catabolism of lipids rather than carbohydrates. The means by which amino acids may be similarly utilised are diverse and are described as appropriate throughout the text. However, we have highlighted glutamine usage in this figure due to its importance. Glutamine may be converted to glutamate by glutaminase. Glutamate may be converted to α-KG by glutamate dehydrogenase (GLUD1) for entry into the TCA cycle, or to aspartate by mitochondrial aspartate aminotransferase (GOT2) which is utilised in cellular redox balancing and TCA cycle anaplerosis, thereby providing both reducing equivalents for OXPHOS and intermediates for the TCA cycle. Reducing equivalents resultant from these myriad processes can deposit electrons into the electron transport chain to facilitate generation of the proton-motive force which drives ATP synthesis.
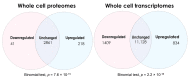
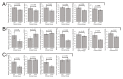
Figure 2
Venn diagrams depicting the numbers of differentially expressed gene products in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome lymphoblasts within the whole-cell proteomics and transcriptomics experiments. Two-sided binomial tests were undertaken with Ho set to p = 0.5 to assess whether the differentially expressed fractions significantly departed from proportions expected by chance. Resulting significance probabilities (p) are indicated.
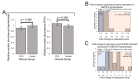
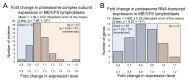
Figure 3
Protein-level expression of glycolytic enzymes is unchanged in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) lymphoblasts. Error bars represent the standard errors of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act as an internal control between experiments in the proteomics work. (A) The expression level of hexokinase and phosphofructokinase is unchanged in whole-cell mass spectrometry proteomics experiments (independent t test). Relative hexokinase (HK) and phosphofructokinase (PFK) abundance was calculated by averaging the mean fold change in the ME/CFS group for each isoenzyme/subunit of the respective enzyme (none of which were statistically significant on their own, threshold p < 0.05). (B) A total of 16 glycolytic enzymes were detected within the whole-cell proteomes of lymphoblasts from ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. There was no significant difference in the differentially expressed proportions of detected glycolytic enzymes (binomial test with Ho set to 0.5) or the magnitude of expression (single-sample t test with Ho m = 1) between ME/CFS and controls. (C) A total of 21 RNA transcripts encoding glycolytic enzymes were detected by RNA sequencing within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Mean fold change was calculated for the ME/CFS group versus the control average for each transcript. The proportion of detected transcripts that were upregulated (binomial test with Ho set to 0.5) was not significant while the average extent of the upregulation (single-sample t test with Ho m = 1) was statistically significant.
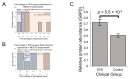
Figure 4
Expression of the pentose phosphate pathway (PPP) is upregulated at the protein level in Myalgic Encephalomyelitis (ME/CFS) lymphoblasts. Error bars represent the standard error of the mean. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments. (A) A total of 7 PPP enzymes were detected within the whole-cell proteomes of lymphoblasts from ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. There was no significant difference in the proportion of upregulated PPP enzymes (binomial test with Ho set to 0.5), but the magnitude of upregulation was significantly elevated in ME/CFS lymphoblasts (single-sample t test with Ho m ≤1 and H1 m>1, p = 0.034). (B) A total of 7 RNA transcripts encoding PPP enzymes were detected by RNA sequencing within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Mean fold change was calculated for the ME/CFS group versus the control average for each transcript. The proportions of reduced or elevated transcripts were not significantly different (binomial test with Ho set to 0.5) nor was the average magnitude of expression (single-sample t test with Ho m = 1). (C) The expression level of G6PD is significantly elevated (t test, p = 5.5 × 10−4) in whole-cell mass spectrometry proteomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments.
Figure 5
Expression of proteins involved in mitochondrial and peroxisomal fatty acid β-oxidation was elevated in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) lymphoblasts. Error bars represent the standard error of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments in the proteomics work. (A) A total of 21 proteins involved in mitochondrial fatty acid β-oxidation and transport were detected within the whole-cell proteomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. The proportion of detected proteins that were upregulated (binomial test with Ho set to 0.5 and H1 being that the upregulated proportion was greater) and the average extent of the upregulation (single-sample t test with Ho ≤ m1 and H1 m>1) were statistically significant. (B) A total of 25 RNA transcripts encoding proteins involved in mitochondrial fatty acid β-oxidation and transport were detected by RNA sequencing within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given transcript in the CFS group divided by the mean abundance in the control group. The proportions of reduced or elevated transcripts were not significantly different (binomial test with Ho p = 0.5) nor was the average magnitude of expression (single-sample t test with Ho m = 1). (C) The expression levels of both subunits of the mitochondrial trifunctional enzyme complex Hydroxyacyl-CoA dehydrogenase/3-keotacyl-CoA thiolase (HADHA and HADHB), very long-chain specific acyl-CoA dehydrogenase (ACADVL), enoyl-CoA hydratase (ECHS1) and electron transfer flavoprotein subunit alpha (ETFA (are significantly elevated in whole-cell mass spectrometry proteomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. (D) The expression of Acyl-CoA oxidase 1 (ACOX1) was significantly elevated in whole-cell mass spectrometry proteomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. ACOX1 expression was not altered at the transcriptional level as measured by whole-cell RNA sequencing transcriptomics. Counts per million mapped reads were calculated for each gene transcript.
Figure 6
Expression of enzymes involved in mitochondrial fatty acid biosynthesis was elevated in the whole-cell transcriptomes but unchanged in the whole-cell proteomes of ME/CFS lymphoblasts. AMPK activity is not significantly elevated in ME/CFS lymphoblasts. Error bars represent the standard error of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments in the proteomics work. (A) A total of 15 proteins involved in fatty acid biosynthesis were detected within the whole-cell proteomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. The proportion of detected proteins that were differentially expressed (binomial test with Ho set to 0.5) and the average extent of any differences (single-sample t test with Ho m = 1) were not statistically significant. (B) A total of 51 RNA transcripts encoding proteins involved in fatty acid biosynthesis were detected by RNA sequencing within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Mean fold change was calculated for the ME/CFS group versus the control average for each transcript. The proportions of reduced or elevated transcripts were not significantly different (binomial test with Ho set to 0.5) nor was the average magnitude of expression (single-sample t test with Ho m = 1). (C) The expression of ACACA and FASN, two key enzymes involved in fatty acid biosynthesis, was not significantly altered in whole-cell proteomics or transcriptomics experiments (independent t test). Relative abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. per million mapped reads were calculated for each gene transcript (D) AMPK activity is not significantly elevated in ME/CFS lymphoblasts. Total ACC levels were unaltered. AMPK activity was determined by measuring the ACC phosphorylation state normalised to total ACC levels in ME/CFS lymphoblasts (n = 28) and healthy controls (n = 24). Each cell line was measured in at least three independent experiments. Fluorescence is expressed in relative terms as each experiment is normalised to an internal control cell line.
Figure 7
Expression of proteins involved in mitochondrial glutamine, BCAA, lysine, tryptophan and phenylalanine utilisation are elevated in ME/CFS lymphoblasts. Error bars represent the standard error of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments in the mass spectrometry proteomics work. (A) Expression of the three enzymes mediating mitochondrial utilisation of glutamate (GLS, GLUD1 and GOT2) were elevated in the whole-cell proteomes and proteomes of ME/CFS lymphoblasts and control lymphoblasts (t test, p < 0.05 in all three cases), while each trended upwards but were not significantly elevated at the transcript level. Relative protein abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. Counts per million mapped reads were calculated for each gene transcript. (B) In ME/CFS lymphoblasts, the expression of BCAT1 is unchanged at the protein and transcript levels, while BCAT2 was unchanged transcriptionally and not detected at the protein level. The levels of BCKDH subunits BCKDHA and BCKDHB are both significantly elevated at the transcriptional and protein levels (t test, p < 0.05), with the exception of BCKDHA transcripts. Relative protein abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual experiments. Counts per million mapped reads were calculated for each gene transcript (C) The expression levels of GCDH and FAH were unchanged at the transcriptional level but elevated at the protein level (t test, p < 0.05) in ME/CFS lymphoblasts. Relative protein abundance was obtained from Intensity-Based Absolute Quantitation values normalised to the control average within the respective individual proteomics experiments. Counts per million mapped reads were calculated for each gene transcript.
Figure 8
Expression of proteasome complexes in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome (ME/CFS) lymphoblasts is upregulated at the protein level but downregulated at the transcript level. Error bars represent the standard error of the mean. RNA sequencing transcriptomics experiment: ME/CFS n = 23, control n = 17. Each cell line was sampled once. Mass spectrometry proteomics experiment: ME/CFS n = 34, control n = 31. Each cell line was sampled once, or twice for a subset of healthy controls arbitrarily selected to act an internal control between experiments in the mass spectrometry proteomics work. (A) A total of 46 proteasome complex subunits were detected within the whole-cell proteomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given protein in the CFS group divided by the mean abundance in the control group. The fraction of detected proteins that were upregulated (binomial test with Ho set to 0.5) and the average extent of the upregulation (single-sample t test with Ho m = 1) were statistically significant. (B) A total of 48 RNA transcripts encoding proteasome complex subunits were detected within the whole-cell transcriptomes of ME/CFS and control lymphoblasts. Fold change refers to the mean abundance of a given transcript in the CFS group divided by the mean abundance in the control group. The fraction of detected transcripts that were downregulated (binomial test with Ho set to 0.5) and the average extent of the downregulation (single-sample t test with Ho m = 1) were statistically significant.
Similar articles
- An Isolated Complex V Inefficiency and Dysregulated Mitochondrial Function in Immortalized Lymphocytes from ME/CFS Patients.
Missailidis D, Annesley SJ, Allan CY, Sanislav O, Lidbury BA, Lewis DP, Fisher PR. Missailidis D, et al. Int J Mol Sci. 2020 Feb 6;21(3):1074. doi: 10.3390/ijms21031074. Int J Mol Sci. 2020. PMID: 32041178 Free PMC article. - Cell-Based Blood Biomarkers for Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.
Missailidis D, Sanislav O, Allan CY, Annesley SJ, Fisher PR. Missailidis D, et al. Int J Mol Sci. 2020 Feb 8;21(3):1142. doi: 10.3390/ijms21031142. Int J Mol Sci. 2020. PMID: 32046336 Free PMC article.
Cited by
- Spotlight on GOT2 in Cancer Metabolism.
Kerk SA, Garcia-Bermudez J, Birsoy K, Sherman MH, Shah YM, Lyssiotis CA. Kerk SA, et al. Onco Targets Ther. 2023 Aug 22;16:695-702. doi: 10.2147/OTT.S382161. eCollection 2023. Onco Targets Ther. 2023. PMID: 37635751 Free PMC article. Review. - Towards an understanding of physical activity-induced post-exertional malaise: Insights into microvascular alterations and immunometabolic interactions in post-COVID condition and myalgic encephalomyelitis/chronic fatigue syndrome.
Haunhorst S, Dudziak D, Scheibenbogen C, Seifert M, Sotzny F, Finke C, Behrends U, Aden K, Schreiber S, Brockmann D, Burggraf P, Bloch W, Ellert C, Ramoji A, Popp J, Reuken P, Walter M, Stallmach A, Puta C. Haunhorst S, et al. Infection. 2024 Sep 6. doi: 10.1007/s15010-024-02386-8. Online ahead of print. Infection. 2024. PMID: 39240417 Review. - Chronic Activation of AMPK Induces Mitochondrial Biogenesis through Differential Phosphorylation and Abundance of Mitochondrial Proteins in Dictyostelium discoideum.
Heidorn-Czarna M, Heidorn HM, Fernando S, Sanislav O, Jarmuszkiewicz W, Mutzel R, Fisher PR. Heidorn-Czarna M, et al. Int J Mol Sci. 2021 Oct 28;22(21):11675. doi: 10.3390/ijms222111675. Int J Mol Sci. 2021. PMID: 34769115 Free PMC article. - Lymphoblastoid Cell Lines as Models to Study Mitochondrial Function in Neurological Disorders.
Annesley SJ, Fisher PR. Annesley SJ, et al. Int J Mol Sci. 2021 Apr 26;22(9):4536. doi: 10.3390/ijms22094536. Int J Mol Sci. 2021. PMID: 33926115 Free PMC article. Review. - Metabolomic Evidence for Peroxisomal Dysfunction in Myalgic Encephalomyelitis/Chronic Fatigue Syndrome.
Che X, Brydges CR, Yu Y, Price A, Joshi S, Roy A, Lee B, Barupal DK, Cheng A, Palmer DM, Levine S, Peterson DL, Vernon SD, Bateman L, Hornig M, Montoya JG, Komaroff AL, Fiehn O, Lipkin WI. Che X, et al. Int J Mol Sci. 2022 Jul 18;23(14):7906. doi: 10.3390/ijms23147906. Int J Mol Sci. 2022. PMID: 35887252 Free PMC article.
References
- Armstrong C.W., McGregor N.R., Lewis D., Butt H., Gooley P.R. Metabolic profiling reveals anomalous energy metabolism and oxidative stress pathways in chronic fatigue syndrome patients. Metabolomics. 2015;11:1626–1639. doi: 10.1007/s11306-015-0816-5. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous