pCysMod: Prediction of Multiple Cysteine Modifications Based on Deep Learning Framework - PubMed (original) (raw)
pCysMod: Prediction of Multiple Cysteine Modifications Based on Deep Learning Framework
Shihua Li et al. Front Cell Dev Biol. 2021.
Abstract
Thiol groups on cysteines can undergo multiple post-translational modifications (PTMs), acting as a molecular switch to maintain redox homeostasis and regulating a series of cell signaling transductions. Identification of sophistical protein cysteine modifications is crucial for dissecting its underlying regulatory mechanism. Instead of a time-consuming and labor-intensive experimental method, various computational methods have attracted intense research interest due to their convenience and low cost. Here, we developed the first comprehensive deep learning based tool pCysMod for multiple protein cysteine modification prediction, including _S_-nitrosylation, _S_-palmitoylation, _S_-sulfenylation, _S_-sulfhydration, and _S_-sulfinylation. Experimentally verified cysteine sites curated from literature and sites collected by other databases and predicting tools were integrated as benchmark dataset. Several protein sequence features were extracted and united into a deep learning model, and the hyperparameters were optimized by particle swarm optimization algorithms. Cross-validations indicated our model showed excellent robustness and outperformed existing tools, which was able to achieve an average AUC of 0.793, 0.807, 0.796, 0.793, and 0.876 for _S_-nitrosylation, _S_-palmitoylation, _S_-sulfenylation, _S_-sulfhydration, and _S_-sulfinylation, demonstrating pCysMod was stable and suitable for protein cysteine modification prediction. Besides, we constructed a comprehensive protein cysteine modification prediction web server based on this model to benefit the researches finding the potential modification sites of their interested proteins, which could be accessed at http://pcysmod.omicsbio.info. This work will undoubtedly greatly promote the study of protein cysteine modification and contribute to clarifying the biological regulation mechanisms of cysteine modification within and among the cells.
Keywords: deep learning; feature extraction; post-translational modifications; prediction; protein cysteine modifications.
Copyright © 2021 Li, Yu, Wu, Zhang, Wang, Zheng, Liu, Wang, Gao and Cheng.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
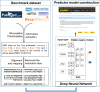
FIGURE 1
An overview of the model.
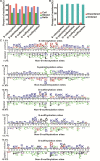
FIGURE 2
The characteristic of cysteine modification sites and proteins. (A) The secondary structure. (B) The disorder information of cysteine modification sites. (C) Preference for amino acids around the cysteine modification sites and non-cysteine modification sites.
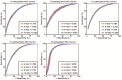
FIGURE 3
The ROC curves and AUCs of 4-, 6-, 8-, and tenfold cross-validations are shown.
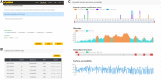
FIGURE 4
The web server of pCysMod. (A) The prediction page. (B) Potential cysteine modification sites. (C) Secondary structure and surface accessibility.
Similar articles
- iCysMod: an integrative database for protein cysteine modifications in eukaryotes.
Wang P, Zhang Q, Li S, Cheng B, Xue H, Wei Z, Shao T, Liu ZX, Cheng H, Wang Z. Wang P, et al. Brief Bioinform. 2021 Sep 2;22(5):bbaa400. doi: 10.1093/bib/bbaa400. Brief Bioinform. 2021. PMID: 33406221 - Deep learning based prediction of species-specific protein S-glutathionylation sites.
Li S, Yu K, Wang D, Zhang Q, Liu ZX, Zhao L, Cheng H. Li S, et al. Biochim Biophys Acta Proteins Proteom. 2020 Jul;1868(7):140422. doi: 10.1016/j.bbapap.2020.140422. Epub 2020 Mar 29. Biochim Biophys Acta Proteins Proteom. 2020. PMID: 32234550 - DeepNitro: Prediction of Protein Nitration and Nitrosylation Sites by Deep Learning.
Xie Y, Luo X, Li Y, Chen L, Ma W, Huang J, Cui J, Zhao Y, Xue Y, Zuo Z, Ren J. Xie Y, et al. Genomics Proteomics Bioinformatics. 2018 Aug;16(4):294-306. doi: 10.1016/j.gpb.2018.04.007. Epub 2018 Sep 27. Genomics Proteomics Bioinformatics. 2018. PMID: 30268931 Free PMC article. - Activity-Based Sensing for Site-Specific Proteomic Analysis of Cysteine Oxidation.
Shi Y, Carroll KS. Shi Y, et al. Acc Chem Res. 2020 Jan 21;53(1):20-31. doi: 10.1021/acs.accounts.9b00562. Epub 2019 Dec 23. Acc Chem Res. 2020. PMID: 31869209 Free PMC article. Review. - GPS-Palm: a deep learning-based graphic presentation system for the prediction of S-palmitoylation sites in proteins.
Ning W, Jiang P, Guo Y, Wang C, Tan X, Zhang W, Peng D, Xue Y. Ning W, et al. Brief Bioinform. 2021 Mar 22;22(2):1836-1847. doi: 10.1093/bib/bbaa038. Brief Bioinform. 2021. PMID: 32248222 Review.
Cited by
- A multienzyme S-nitrosylation cascade regulates cholesterol homeostasis.
Stomberski CT, Venetos NM, Zhou HL, Qian Z, Collison BR, Field SJ, Premont RT, Stamler JS. Stomberski CT, et al. Cell Rep. 2022 Oct 25;41(4):111538. doi: 10.1016/j.celrep.2022.111538. Cell Rep. 2022. PMID: 36288700 Free PMC article. - S-Nitrosation of E3 Ubiquitin Ligase Complex Components Regulates Hormonal Signalings in Arabidopsis.
Terrile MC, Tebez NM, Colman SL, Mateos JL, Morato-López E, Sánchez-López N, Izquierdo-Álvarez A, Marina A, Calderón Villalobos LIA, Estelle M, Martínez-Ruiz A, Fiol DF, Casalongué CA, Iglesias MJ. Terrile MC, et al. Front Plant Sci. 2022 Feb 4;12:794582. doi: 10.3389/fpls.2021.794582. eCollection 2021. Front Plant Sci. 2022. PMID: 35185952 Free PMC article. - Sul-BertGRU: an ensemble deep learning method integrating information entropy-enhanced BERT and directional multi-GRU for S-sulfhydration sites prediction.
Wei X, Ning Q, Che K, Liu Z, Li H, Guo S. Wei X, et al. Bioinformatics. 2025 Mar 4;41(3):btaf078. doi: 10.1093/bioinformatics/btaf078. Bioinformatics. 2025. PMID: 39977366 Free PMC article. - PPSNO: A Feature-Rich SNO Sites Predictor by Stacking Ensemble Strategy from Protein Sequence-Derived Information.
Zhu L, Wang L, Yang Z, Xu P, Yang S. Zhu L, et al. Interdiscip Sci. 2024 Mar;16(1):192-217. doi: 10.1007/s12539-023-00595-7. Epub 2024 Jan 11. Interdiscip Sci. 2024. PMID: 38206557 - Genome-Wide In Silico Analysis of 1-Aminocyclopropane-1-carboxylate oxidase (ACO) Gene Family in Rice (Oryza sativa L.).
Xia J, Qiu Y, Li W, Zhang Y, Liu L, Wang Y, Mou W, Xue D. Xia J, et al. Plants (Basel). 2024 Dec 13;13(24):3490. doi: 10.3390/plants13243490. Plants (Basel). 2024. PMID: 39771188 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous