A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids - PubMed (original) (raw)
A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids
Jürgen F H Strassert et al. Nat Commun. 2021.
Erratum in
- Author Correction: A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids.
Strassert JFH, Irisarri I, Williams TA, Burki F. Strassert JFH, et al. Nat Commun. 2021 Jun 7;12(1):3574. doi: 10.1038/s41467-021-23847-w. Nat Commun. 2021. PMID: 34099732 Free PMC article. No abstract available.
Abstract
In modern oceans, eukaryotic phytoplankton is dominated by lineages with red algal-derived plastids such as diatoms, dinoflagellates, and coccolithophores. Despite the ecological importance of these groups and many others representing a huge diversity of forms and lifestyles, we still lack a comprehensive understanding of their evolution and how they obtained their plastids. New hypotheses have emerged to explain the acquisition of red algal-derived plastids by serial endosymbiosis, but the chronology of these putative independent plastid acquisitions remains untested. Here, we establish a timeframe for the origin of red algal-derived plastids under scenarios of serial endosymbiosis, using Bayesian molecular clock analyses applied on a phylogenomic dataset with broad sampling of eukaryote diversity. We find that the hypotheses of serial endosymbiosis are chronologically possible, as the stem lineages of all red plastid-containing groups overlap in time. This period in the Meso- and Neoproterozoic Eras set the stage for the later expansion to dominance of red algal-derived primary production in the contemporary oceans, which profoundly altered the global geochemical and ecological conditions of the Earth.
Conflict of interest statement
The authors declare no competing interests.
Figures
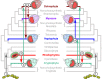
Fig. 1. Serial plastid endosymbioses models as proposed by Stiller et al. (left) and Bodył et al. (right).
The tree topology shown here is based on the results obtained in our study. Further models have been suggested but are not compatible with this topology,,. Numbers denote the level of endosymbiosis events. Note, Myzozoa were not included by Stiller et al. and the dashed line indicates engulfment of an ochrophyte by the common ancestor of Myzozoa as suggested by Sevcikova et al..
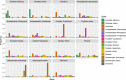
Fig. 2. Test for endosymbiotic gene transfers in red plastid-containing lineages based on the analysis of 320 single-gene ML trees.
For each clade in the ML tree to which a single taxonomic label could be assigned, relative frequencies with which all other clades in the tree were recovered as the closest sister group are given (for details, see ‘Methods’).
Fig. 3. Time-calibrated phylogeny of extant eukaryotes.
Divergence times were inferred with MCMCTree under an autocorrelated relaxed clock model and 33 fossil calibration points as soft-bound uniform priors (Table 1). The tree topology was reconstructed using IQ-TREE under the LG + C60 + G + F model and a constrained tree search following the OTU-reduced Bayesian CAT + GTR + G topology (Supplementary Fig. 4). Approximate likelihood calculations on the 320 gene concatenation under LG + G and a birth–death tree prior were used. Bars at nodes are 95% HPD. Bars corresponding to the first and last common ancestors of extant red plastid-donating and -containing lineages are highlighted in red and their stems are shaded as indicated. Crowns denote the common ancestors of the extant members of these groups. An absolute time scale in Ma and a geological time scale are shown. The tree depicted here was rooted on Amorphea. An equivalent time-calibrated tree rooted on Excavata is shown in Supplementary Fig. 7. Cry Cryptophyta, Hap Haptophyta, Myz Myzozoa, Och Ochrophyta, Rho Rhodophytina.
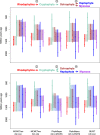
Fig. 4. Summary of inferred timeframes for the spread of complex red plastids using different software and models.
Vertical lines correspond to the lower and upper 95% HPD intervals from the nodes defining the branch of interest and dots indicate their posterior mean divergences. Faded boxes represent the temporal windows for the secondary endosymbioses, constrained by the 95% HPDs and tree topology under the two proposed symbiotic scenarios. Numbers at arrows denote the level of endosymbiosis events (compare with Fig. 1). AC autocorrelated clock model, UC uncorrelated clock model.
Similar articles
- A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids.
Janouskovec J, Horák A, Oborník M, Lukes J, Keeling PJ. Janouskovec J, et al. Proc Natl Acad Sci U S A. 2010 Jun 15;107(24):10949-54. doi: 10.1073/pnas.1003335107. Epub 2010 Jun 1. Proc Natl Acad Sci U S A. 2010. PMID: 20534454 Free PMC article. - Chromera velia, endosymbioses and the rhodoplex hypothesis--plastid evolution in cryptophytes, alveolates, stramenopiles, and haptophytes (CASH lineages).
Petersen J, Ludewig AK, Michael V, Bunk B, Jarek M, Baurain D, Brinkmann H. Petersen J, et al. Genome Biol Evol. 2014 Mar;6(3):666-84. doi: 10.1093/gbe/evu043. Genome Biol Evol. 2014. PMID: 24572015 Free PMC article. - Algal genes in the closest relatives of animals.
Sun G, Yang Z, Ishwar A, Huang J. Sun G, et al. Mol Biol Evol. 2010 Dec;27(12):2879-89. doi: 10.1093/molbev/msq175. Epub 2010 Jul 13. Mol Biol Evol. 2010. PMID: 20627874 - The endosymbiotic origin, diversification and fate of plastids.
Keeling PJ. Keeling PJ. Philos Trans R Soc Lond B Biol Sci. 2010 Mar 12;365(1541):729-48. doi: 10.1098/rstb.2009.0103. Philos Trans R Soc Lond B Biol Sci. 2010. PMID: 20124341 Free PMC article. Review. - A new scenario of plastid evolution: plastid primary endosymbiosis before the divergence of the "Plantae," emended.
Nozaki H. Nozaki H. J Plant Res. 2005 Aug;118(4):247-55. doi: 10.1007/s10265-005-0219-1. Epub 2005 Jul 20. J Plant Res. 2005. PMID: 16032387 Review.
Cited by
- Early Diversification of Membrane Intrinsic Proteins (MIPs) in Eukaryotes.
Irisarri I, Lorente-Martínez H, Strassert JFH, Agorreta A, Zardoya R, San Mauro D, de Vries J. Irisarri I, et al. Genome Biol Evol. 2024 Aug 5;16(8):evae164. doi: 10.1093/gbe/evae164. Genome Biol Evol. 2024. PMID: 39058319 Free PMC article. - Amoebozoan testate amoebae illuminate the diversity of heterotrophs and the complexity of ecosystems throughout geological time.
Porfirio-Sousa AL, Tice AK, Morais L, Ribeiro GM, Blandenier Q, Dumack K, Eglit Y, Fry NW, Gomes E Souza MB, Henderson TC, Kleitz-Singleton F, Singer D, Brown MW, Lahr DJG. Porfirio-Sousa AL, et al. Proc Natl Acad Sci U S A. 2024 Jul 23;121(30):e2319628121. doi: 10.1073/pnas.2319628121. Epub 2024 Jul 16. Proc Natl Acad Sci U S A. 2024. PMID: 39012821 Free PMC article. - A unique flavoenzyme operates in ubiquinone biosynthesis in photosynthesis-related eukaryotes.
Xu JJ, Zhang XF, Jiang Y, Fan H, Li JX, Li CY, Zhao Q, Yang L, Hu YH, Martin C, Chen XY. Xu JJ, et al. Sci Adv. 2021 Dec 10;7(50):eabl3594. doi: 10.1126/sciadv.abl3594. Epub 2021 Dec 8. Sci Adv. 2021. PMID: 34878842 Free PMC article. - IRplus: An Augmented Tool to Detect Inverted Repeats in Plastid Genomes.
Díez Menéndez C, Poczai P, Williams B, Myllys L, Amiryousefi A. Díez Menéndez C, et al. Genome Biol Evol. 2023 Oct 6;15(10):evad177. doi: 10.1093/gbe/evad177. Genome Biol Evol. 2023. PMID: 37793175 Free PMC article. - Phylogeny and evolution of streptophyte algae.
Bierenbroodspot MJ, Pröschold T, Fürst-Jansen JMR, de Vries S, Irisarri I, Darienko T, de Vries J. Bierenbroodspot MJ, et al. Ann Bot. 2024 Aug 22;134(3):385-400. doi: 10.1093/aob/mcae091. Ann Bot. 2024. PMID: 38832756 Free PMC article. Review.
References
- Holland HD. Volcanic gases, black smokers, and the great oxidation event. Geochim. Cosmochim. Acta. 2002;66:3811–3826.
- Irisarri, I., Strassert, J. F. H. & Burki, F. Phylogenomic insights into the origin of primary plastids. bioRxiv10.1101/2020.08.03.231043 (2020). - PubMed
- Burki, F. in Secondary Endosymbioses Vol. 84 (ed. Hirakawa, Y. B. T.-A. in B. R.) 1–30 (Academic, 2017).
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources