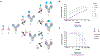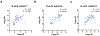Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection - PubMed (original) (raw)
. 2021 Aug;39(8):928-935.
doi: 10.1038/s41587-021-00878-8. Epub 2021 Mar 25.
Xin X Zhou # 1, James R Byrnes 1, Alexander J Martinko 2, Irene Lui 1, Katarina Pance 1, Shion A Lim 1, Jeff E Glasgow 1, Anum A Glasgow 3, Keirstinne Turcios 4, Nikita S Iyer 4, Leonel Torres 4 5, Michael J Peluso 5, Timothy J Henrich 4, Taia T Wang 6 7, Cristina M Tato 6, Kevin K Leung 1, Bryan Greenhouse 4 5, James A Wells 8 9 10
Affiliations
- PMID: 33767397
- PMCID: PMC8355051
- DOI: 10.1038/s41587-021-00878-8
Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection
Susanna K Elledge et al. Nat Biotechnol. 2021 Aug.
Abstract
Current serology tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibodies mainly take the form of enzyme-linked immunosorbent assays, chemiluminescent microparticle immunoassays or lateral flow assays, which are either laborious, expensive or lacking sufficient sensitivity and scalability. Here we present the development and validation of a rapid, low-cost, solution-based assay to detect antibodies in serum, plasma, whole blood and to a lesser extent saliva, using rationally designed split luciferase antibody biosensors. This new assay, which generates quantitative results in 30 min, substantially reduces the complexity and improves the scalability of coronavirus disease 2019 (COVID-19) antibody tests. This assay is well-suited for point-of-care, broad population testing, and applications in low-resource settings, for monitoring host humoral responses to vaccination or viral infection.
© 2021. The Author(s), under exclusive licence to Springer Nature America, Inc.
Figures
Extended Data Fig. 1 ∣. Design and characterization of S sensors.
a, Annotated depiction of the SARS-CoV-2 Spike protein. The S sensors were developed using only the S-RBD domain (aa 328–533, PDB: 6W41) shown in pink. b, Structure of the S-RBD domain shows the N and C termini locate in close proximity. c, d, Modeling of c, ACE2-competitive antibody C105 (PDB: 6XCN) binding to S-RBD-SmBiT/LgBiT sensors, and d, CR3022 (PDB: 6W41) binding to S-RBD-SmBiT/LgBiT sensors. Modeling and distance measurements were performed with PDB 6XCN, 6W41, 1N8Z, 5IBO and 5D6D in PyMOL. e, Yield of the 5 Spike-NanoBiT sensor fusions. The Spike LgBiT sensors were made with 5aa, 15aa, and 25aa Glycine-Serine (GS) linkers (L5, L15 and L25). The Spike SmBiT sensors were made with 15aa, and 25aa GS linkers (S15 and S25). Because the N and C termini of the S-RBD domain locate in close proximity, only fusions to the C termini of S-RBD were constructed. f, The S sensors are most sensitive at 1 nM for detecting CR3022 in solution compared to higher or lower sensor concentrations. Two technical replicates are plotted from n=1 individual experiment. Lines connecting the means of the samples are plotted. g, S sensors with varied linker lengths resulted in very similar signal strength in detecting CR3022. Two technical replicates are plotted from n=1 individual experiment. Lines connecting the means of the samples are plotted.
Extended Data Fig. 2 ∣. The biosensors are more sensitive to high-affinity binders.
The ACE2-Fc variant which binds 10-fold tighter to S-RBD generated ~3-fold higher signal at 10 nM protein concentration comparing to WT ACE2-Fc. Two technical replicates are plotted from n=1 individual experiment. Lines connecting the means of the samples are plotted.
Extended Data Fig. 3 ∣. ODE models predict a linear, dose-dependent response and KD dependence of the luminescence signal.
a, Antibody (C) and sensor components (A and B) are in thermodynamic equilibrium with enzymatically inactive (D, E, G, and I) and active (H) sensor bound species. b, At 1 nM starting concentration of sensor ([A] and [B]), spLUC assays are predicted to generate signals linearly correlated to a broad range of antibody concentrations ([Ab]). Signal is predicted to be insensitive to antibody concentrations for antibodies with high affinity for the sensor (≤ 1nM), but weaker affinity antibodies (KD > 1 nM) will result in significantly lower levels of reconstituted enzyme. c, At KD values equivalent or higher than the sensor concentrations, the spLUC signals are predicted to drop significantly.
Extended Data Fig. 4 ∣. Design and characterization of N sensors.
a, Annotated depiction of the SARS-CoV-2 Nucleocapsid protein (protein N). All N protein fusions designed included the RNA binding domain (aa 44–180, N-RBD) and excluded the dimerization domain (aa 257–419). b, Structure of the N-RBD domain shows the N and C termini locate far from each other and fusion of the split enzyme fragments to N or C termini may result in different detection sensitivity (PDB: 6YI3). c, Yield of the six N protein-NanoBiT sensor fusions. d, The N-terminal N sensor pair (LN + SN, 44–257) was less sensitive than the LC + SC (44–180) and LC2 + SC2 (44–257) C terminal N sensor pairs when the assay was performed on a rabbit polyclonal anti-N protein antibody (Sino Biological, Cat#: 40588-T62-50). Two technical replicates are plotted from n=1 individual experiment. Lines connecting the means of the samples are plotted. e, Additionally, only patient 6 and 8 showed signals above controls in the serological assay performed with LN + SN sensors, while all four patients showed signals with the LC + SC sensors. Two technical replicates are plotted from n = 1 independent experiment. Lines connecting the means of the samples are plotted.
Extended Data Fig. 5 ∣. Individual cohorts show good correlation between S and N sensors.
Each cohort shows robust correlation with R = 0.59, 0.87, and 0.73 for a, cohort 1 (56 samples), b, cohort 2 (47 samples), and c, cohort 3 (87 samples), respectively. R values and P values (two-tail) of a non-parametric Spearman correlation analysis are labeled in the graphs. Lines represent linear regression. For all graphs, dots represent the average of two technical replicates from n=1 independent experiment.
Extended Data Fig. 6 ∣. Comparison of the ELISA and the spLUC results.
a, Signals from the S sensor spLUC assay (cohort 1, 57 samples) correlate very well with S-RBD ELISA anti-Fab signals (R = 0.91), moderately well with anti-IgG signals (R = 0.43), and poorly with anti-IgM signals for cohort 1 (R = −0.066). Line represents linear regression. b, Signals from the S sensor spLUC assay (cohort 2, 40 samples with anti-Fab detection, 47 samples with anti-IgM/IgG detection) correlate very well with S-RBD ELISA anti-Fab signals (R = 0.84) and with anti-IgG signals (R = 0.86), but poorly with anti-IgM signals for cohort 1 (R = 0.29). Line represents linear regression. c, Signals from the S sensor (cohort 3, 87 samples) correlate well with S-RBD ELISA anti-IgG signals (R = 0.88). For a-c, R values and P values (two-tail) of a non-parametric Spearman correlation analysis are labeled in the graphs. Lines represent linear regression. d, e, f, The seronegative samples in the anti-S spLUC assay also showed low anti-Fab or anti-IgG signals in ELISA serology tests for d, cohort 1 (negative samples (9), positive samples (48)), e, cohort 2, (negative samples (5), positive samples (35)), and f, cohort 3, (negative samples (3), positive samples (53)). Horizontal lines represent the median value. For all graphs, dots represent the average of two technical replicates from n=1 independent experiment.
Extended Data Fig. 7 ∣. Inter-assay, Inter-day and Intra-day variability of spLUC assay.
a, 46 plasma samples were assayed a total of five times in three independent experiments over two days for each sensor. All five replicates are plotted on the graph, with the bar representing the average. The dotted line represents the cutoff values for positive and negative samples for the S and N sensors. b, The coefficient of variation was calculated for intra-assay, intra-day, and inter-day variability. Coefficient of variation is calculated as ratio of standard deviation to the average value.
Extended Data Fig. 8 ∣. Simulated robotics-assisted spLUC assay.
a, Serum sample transfer to an assay plate using Biomek Fx Automated Workstation in ~2 minutes. b, Robotics-assisted dispensing and luminescence reading for one iteration of 96 assays takes ~35 minutes. c, Simulated run for 40 iterations (3840 assays) can be completed in 3 hours. Gantt chart generated by simulated run using Thermo Momentum software.
Extended Data Fig. 9 ∣. S and N sensors are functional after lyophilization.
a, Both the S and the N sensors can survive lyophilization. The majority of proteins (70–100%) can be reconstituted after lyophilization. The lyophilized S sensors lost 50% of signal. The lyophilized N sensors remain 100% active. b, The lyophilized S sensors detected CR3022 at ~50% signal strength compared to fresh sensors. Three technical replicates are plotted from n=1 independent experiment. Lines connecting the means of the samples are plotted. c, The lyophilized N sensors detected antibodies from patient sera at similar signal strength compared to fresh sensors. Two technical replicates are plotted from n=1 independent experiment. The bars represent the mean. d, Vacuum dried substrate and substrate stored at −20°C (fresh) behave similarly when detecting recombinant CR3022 with S sensors. Lyophilized dilution buffer and dilution buffer stored at −20 °C (fresh) also showed similar signal. Three technical replicates are plotted from n=1 independent experiment. Lines connecting the means of the samples are plotted.
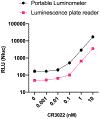
Extended Data Fig. 10 ∣. Portable luminometer.
The spLUC assay is also amenable to detection with a Berthold portable luminometer. The handheld luminometer showed similar sensitivity of recombinant CR3022 with S sensors compared to the plate reader. Due to the tube format of the handheld luminometer, the sample volume was doubled and thus the overall signals are higher than for the plate reader samples, but similar sensitivity is maintained. Two technical replicates are plotted from n=1 independent experiment. Lines connecting the means of the samples are plotted.
Fig. 1 ∣. Engineering luminescent biosensors for rapid and quantitative detection of SARS-CoV-2 antibodies.
a, Schematic of the solution-based serology assay. Patient antibodies are incubated with SARS-CoV-2 S or N proteins fused to LgBiT/SmBiT. For the population of antibodies with one arm bound to the LgBiT sensor and the other arm bound to the SmBiT sensor, the NanoBiT luciferase enzyme is reconstituted and, thus, can produce active luciferase signal. b, Dose-dependent spLUC signals for the recombinant anti-S-RBD antibody C004 in PBST + 10% FBS. Two technical replicates are plotted from n = 1 independent experiment. c, Dose-dependent spLUC signals for an anti-N-RBD antibody (Sino Biological, cat. no. 40588-T62–50) in PBST + 8% FBS. Two technical replicates are plotted from n = 1 independent experiment. d, Comparison of assay procedures between the ELISA and the spLUC assay. While ELISA takes more than 2 h and involves multiple wash and incubation steps, the spLUC solution-based assay is simply completed in 30 min or less without the need for wash steps. e, The S (L15 + S25) sensors are able to detect antibodies in 5/5 recovered patients with COVID-19. At all dilutions tested, all five patients generated signal above the background signal of two control serum samples collected before the pandemic. Each dot represents a technical replicate. n = 2 independent experiments with three replicates each are plotted for all samples, except for those of Patient 1, Patient 7 and Control 2, which have n = 1 independent experiment plotted owing to limited reagents. f, The N (LC + SC) sensors are able to detect antibodies in 4/4 recovered patients with COVID-19. At all dilutions of serum tested, all four patients generated signal above the background signal of two control serum samples collected before the pandemic. Two technical replicates are plotted from n = 1 independent experiment. g, Patient antibodies for SARS-CoV-2 have various epitopes on the S-RBD (red). C004 and C105 have ACE2-competitive epitopes, whereas C135 and CR3022 (blue) have non-ACE2-competitive epitopes. h, S sensors can detect patient antibodies of various epitopes with similar sensitivity. C004, C105, C135 and CR3022 patient antibodies were incubated with the S sensors at ten-fold antibody dilutions from 10 nM to 0.001 nM. The average of three technical replicates from n = 2 independent experiments are plotted. i, Schematic of antibody epitope competition assay with patient serum samples. Direct signal is compared to signal generated in the presence of the pre-incubated 1μM Fab +1 nM sensor. j, Competition assay performed with C135 Fab on 12 outpatient sera samples and recombinant C135 IgG protein. Samples were incubated with either no Fab (blue) or C135 Fab (off-white). Sera 7, 42 and 98 showed more than 50% decreases in luminescence signal, suggesting the presence of antibodies with the C135 epitope. Each dot represents a technical replicate from n = 1 independent experiment. The center of the bar represents the mean of the measurements. Direct signals (–Fab) were measured in technical duplicates, and competition signals (+C135 Fab) samples were done in technical triplicate. For b, c, e, f and h, the center of the line represents the mean of all measurements. Lines connecting the means of the samples are plotted. RLU, relative luminescence unit; RT, room temperature; NFM, non-fat milk; O/N, overnight.
Fig. 2 ∣. Characterization of outpatient and inpatient serum samples using the spLUC test.
Cohort 1: samples drawn during the convalescent phase of an outpatient group; Cohort 2: samples drawn during the acute phase or the convalescent phase of a hospitalized group; and Cohort 3: samples drawn during the convalescent phase of a mixed inpatient and outpatient group. A 10-base logarithmic scale conversion was applied to all the solution assay signals for the correlation analysis, unless otherwise specified. a, SpLUC assay tested on expanded COVID-19 patient cohorts with S sensors at 1:12.5 serum dilution. Dots represent the average between two technical duplicates. Lines represent median values. The inpatient samples showed significantly higher antibody titers than the outpatient cohorts. Sample sizes are as indicated in parentheses: Control (56), Cohort 1 (57), Cohort 2 (55), Cohort 3 outpatient (47) and Cohort 3 inpatient (9). b, SpLUC assay tested on expanded COVID-19 patient cohorts with N sensors at 1:12.5 serum dilution. The inpatient samples showed significantly higher antibody titers than the outpatient cohorts. Sample sizes are as indicated in parentheses: Control (120), Cohort 1 (56), Cohort 2 (47), Cohort 3 outpatient (47) and Cohort 3 inpatient (9). c, A positive correlation (R = 0.78) was observed between S sensor signal and N sensor signal in the three cohort samples. All cohorts individually presented a similar trend (Extended Data Fig. 5). The line represents linear regression. In total, 159 patient samples are plotted. d, Correlation of spLUC signals (Cohort 1) to neutralization efficiency. S sensor signal (blue) and N sensor signal (purple) are plotted against 50% maximal neutralization titer (NT50). Both show positive correlation (R = 0.76 for S and NT50 and R = 0.62 for N and NT50). Fifty-seven patient samples are plotted for the S sensor, and 56 samples are plotted for the N sensor. e, Inpatients show significantly higher signal than outpatients in all three cohorts (P < 0.0001). Sample sizes are as indicated in parentheses: S-outpatient (104), S-inpatient (64), N-outpatient (103) and N-inpatient (56). f, Patients from Cohort 1 who reported higher disease severity (6–10 versus 1–5) had higher antibody titer for both S and N sensors, and the difference for N sensors is statistically significant (P = 0.0049). Sample sizes are as indicated in parentheses: S-score 1–5 (24), S-score 6–9 (33), N-score 1–5 (24) and N-score 6–9 (32). g, Higher overall antibody titers were observed in patients who reported fever compared to no-fever patients for Cohort 3. This difference was statistically significant for the S sensors (P = 0.0011) but not for the N sensors. Seventeen patient samples associated with no fever and 38 patient samples associated with a fever are plotted. h, Slightly higher overall antibody titers were observed in females than in males for Cohort 3, although the differences were not statistically significant. There is a similar trend for Cohort 1 (Supplementary Fig. 3a). The difference was more obvious for S sensors. Twenty-five female patient samples and 30 male patient samples are plotted. i, For Cohort 3, there is a slightly higher level of antibodies in the 60–85 age group compared to 19–39 and 40–59. There is a similar trend for Cohort 1 (Supplementary Fig. 3b). The differences were not statistically significant. Sample sizes are as indicated in parentheses: age 19–39 (23), age 40–59 (25) and age 60–85 (8). For a and b, P values (two-tailed) of a Kruskal–Wallis test with Dunn’s multiple comparison post hoc testing to compare each clinical cohort with the healthy control group are indicated. For c and d, R values and P values (two-tailed) of a non-parametric Spearman correlation analysis are labeled in the graphs. For e–i, an unpaired Mann–Whitney test is performed, and P values (two-tailed) for each comparison are labeled on top of the datasets. For all panels, dots represent the average of two technical replicates from n = 1 independent experiment. For a and b and e–i, horizonal lines represent median values. For c and d, lines represent linear regression. RLU, relative luminescence unit.
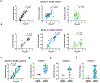
Fig. 3 ∣. Adapting the assay for whole blood and saliva sample types.
a, spLUC assays can be accomplished in as few as 5 min. CR3022 (10 nM) was incubated with S sensors for 5, 10, 15 or 20 min. Luciferase substrates were then added and incubated with the reaction mix for 0, 2, 4, 6, 8 or 10 min. All reactions showed bright luminescence signal. Each dot represents the average of two technical duplicates from n = 1 independent experiment. Lines connecting the means of the samples are plotted. b, The spLUC assay is compatible with whole blood samples and shows similar signal in the corresponding plasma samples with both fresh and lyophilized sensors. Each dot represents the average of two technical replicates from n = 1 independent experiment. A non-parametric Spearman correlation analysis was performed, and R = 0.94 was observed for S sensors, and R = 1 and 0.98 were observed for N sensor fresh and lyophilized sensors, respectively. c, Anti-S antibodies were detected in saliva samples with moderate sensitivity (33/42, 79%). The signals from saliva samples positively correlated with corresponding serum samples. Each dot represents the average of two technical replicates from n = 1 independent experiment. A non-parametric Spearman correlation analysis was performed, and the R value (0.66) and P value (<0.0001, two-tailed) are labeled in the graphs. The line represents linear regression.
Update of
- Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection.
Elledge SK, Zhou XX, Byrnes JR, Martinko AJ, Lui I, Pance K, Lim SA, Glasgow JE, Glasgow AA, Turcios K, Iyer N, Torres L, Peluso MJ, Henrich TJ, Wang TT, Tato CM, Leung KK, Greenhouse B, Wells JA. Elledge SK, et al. medRxiv [Preprint]. 2020 Aug 21:2020.08.17.20176925. doi: 10.1101/2020.08.17.20176925. medRxiv. 2020. PMID: 32839788 Free PMC article. Updated. Preprint.
Similar articles
- Engineering luminescent biosensors for point-of-care SARS-CoV-2 antibody detection.
Elledge SK, Zhou XX, Byrnes JR, Martinko AJ, Lui I, Pance K, Lim SA, Glasgow JE, Glasgow AA, Turcios K, Iyer N, Torres L, Peluso MJ, Henrich TJ, Wang TT, Tato CM, Leung KK, Greenhouse B, Wells JA. Elledge SK, et al. medRxiv [Preprint]. 2020 Aug 21:2020.08.17.20176925. doi: 10.1101/2020.08.17.20176925. medRxiv. 2020. PMID: 32839788 Free PMC article. Updated. Preprint. - Serological assays and host antibody detection in coronavirus-related disease diagnosis.
Dowlatshahi S, Shabani E, Abdekhodaie MJ. Dowlatshahi S, et al. Arch Virol. 2021 Mar;166(3):715-731. doi: 10.1007/s00705-020-04874-2. Epub 2021 Jan 25. Arch Virol. 2021. PMID: 33492524 Free PMC article. Review. - A rapid, point-of-care red blood cell agglutination assay detecting antibodies against SARS-CoV-2.
Kruse RL, Huang Y, Smetana H, Gehrie EA, Amukele TK, Tobian AAR, Mostafa HH, Wang ZZ. Kruse RL, et al. Biochem Biophys Res Commun. 2021 May 14;553:165-171. doi: 10.1016/j.bbrc.2021.03.016. Epub 2021 Mar 15. Biochem Biophys Res Commun. 2021. PMID: 33773139 Free PMC article. - SARS-CoV-2 Antibody Rapid Tests: Valuable Epidemiological Tools in Challenging Settings.
Saluzzo F, Mantegani P, Poletti De Chaurand V, Quaresima V, Cugnata F, Di Serio C, Macé A, De Vos M, Sacks JA, Cirillo DM. Saluzzo F, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0025021. doi: 10.1128/Spectrum.00250-21. Epub 2021 Sep 22. Microbiol Spectr. 2021. PMID: 34549999 Free PMC article. - A comparative review of immunoassays for COVID-19 detection.
Mohit E, Rostami Z, Vahidi H. Mohit E, et al. Expert Rev Clin Immunol. 2021 Jun;17(6):573-599. doi: 10.1080/1744666X.2021.1908886. Expert Rev Clin Immunol. 2021. PMID: 33787412 Review.
Cited by
- Multiplexed bioluminescence imaging with a substrate unmixing platform.
Brennan CK, Yao Z, Ionkina AA, Rathbun CM, Sathishkumar B, Prescher JA. Brennan CK, et al. Cell Chem Biol. 2022 Nov 17;29(11):1649-1660.e4. doi: 10.1016/j.chembiol.2022.10.004. Epub 2022 Oct 24. Cell Chem Biol. 2022. PMID: 36283402 Free PMC article. - Airline Point-of-Care System on Seat Belt for Hybrid Physiological Signal Monitoring.
Ji X, Rao Z, Zhang W, Liu C, Wang Z, Zhang S, Zhang B, Hu M, Servati P, Xiao X. Ji X, et al. Micromachines (Basel). 2022 Nov 1;13(11):1880. doi: 10.3390/mi13111880. Micromachines (Basel). 2022. PMID: 36363901 Free PMC article. - Multivalent designed proteins neutralize SARS-CoV-2 variants of concern and confer protection against infection in mice.
Hunt AC, Case JB, Park YJ, Cao L, Wu K, Walls AC, Liu Z, Bowen JE, Yeh HW, Saini S, Helms L, Zhao YT, Hsiang TY, Starr TN, Goreshnik I, Kozodoy L, Carter L, Ravichandran R, Green LB, Matochko WL, Thomson CA, Vögeli B, Krüger A, VanBlargan LA, Chen RE, Ying B, Bailey AL, Kafai NM, Boyken SE, Ljubetič A, Edman N, Ueda G, Chow CM, Johnson M, Addetia A, Navarro MJ, Panpradist N, Gale M Jr, Freedman BS, Bloom JD, Ruohola-Baker H, Whelan SPJ, Stewart L, Diamond MS, Veesler D, Jewett MC, Baker D. Hunt AC, et al. Sci Transl Med. 2022 May 25;14(646):eabn1252. doi: 10.1126/scitranslmed.abn1252. Epub 2022 May 25. Sci Transl Med. 2022. PMID: 35412328 Free PMC article. - Advancements in Detection Approaches of Severe Acute Respiratory Syndrome Coronavirus 2.
Hossain MAM, Uddin SMK, Hashem A, Mamun MA, Sagadevan S, Johan MR. Hossain MAM, et al. Malays J Med Sci. 2022 Dec;29(6):15-33. doi: 10.21315/mjms2022.29.6.3. Epub 2022 Dec 22. Malays J Med Sci. 2022. PMID: 36818907 Free PMC article. Review. - Rapid biosensing SARS-CoV-2 antibodies in vaccinated healthy donors.
Bian S, Shang M, Sawan M. Bian S, et al. Biosens Bioelectron. 2022 May 15;204:114054. doi: 10.1016/j.bios.2022.114054. Epub 2022 Feb 3. Biosens Bioelectron. 2022. PMID: 35151002 Free PMC article.
References
- Krammer F & Simon V Serology assays to manage COVID-19. Science 368, 1060–1061 (2020). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R35 GM122451/GM/NIGMS NIH HHS/United States
- K24 AI144048/AI/NIAID NIH HHS/United States
- T32 AI060530/AI/NIAID NIH HHS/United States
- R00 GM135529/GM/NIGMS NIH HHS/United States
- U19 AI111825/AI/NIAID NIH HHS/United States
- R01 AI139119/AI/NIAID NIH HHS/United States
- F32 CA239417/CA/NCI NIH HHS/United States
- UL1 TR001866/TR/NCATS NIH HHS/United States
- K99 EB030587/EB/NIBIB NIH HHS/United States
- K99 GM135529/GM/NIGMS NIH HHS/United States
- R01 AI141003/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous