Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease - PubMed (original) (raw)
Observational Study
. 2021 Jul;64(7):1504-1515.
doi: 10.1007/s00125-021-05444-0. Epub 2021 Apr 2.
Girish N Nadkarni 2, Fergus Fleming 3 4, James R McCullough 3 4, Patricia Connolly 3 4, Gohar Mosoyan 2, Fadi El Salem 5, Michael W Kattan 6, Joseph A Vassalotti 2, Barbara Murphy 2, Michael J Donovan 5, Steven G Coca 2, Scott M Damrauer 7
Affiliations
- PMID: 33797560
- PMCID: PMC8187208
- DOI: 10.1007/s00125-021-05444-0
Observational Study
Derivation and validation of a machine learning risk score using biomarker and electronic patient data to predict progression of diabetic kidney disease
Lili Chan et al. Diabetologia. 2021 Jul.
Abstract
Aim: Predicting progression in diabetic kidney disease (DKD) is critical to improving outcomes. We sought to develop/validate a machine-learned, prognostic risk score (KidneyIntelX™) combining electronic health records (EHR) and biomarkers.
Methods: This is an observational cohort study of patients with prevalent DKD/banked plasma from two EHR-linked biobanks. A random forest model was trained, and performance (AUC, positive and negative predictive values [PPV/NPV], and net reclassification index [NRI]) was compared with that of a clinical model and Kidney Disease: Improving Global Outcomes (KDIGO) categories for predicting a composite outcome of eGFR decline of ≥5 ml/min per year, ≥40% sustained decline, or kidney failure within 5 years.
Results: In 1146 patients, the median age was 63 years, 51% were female, the baseline eGFR was 54 ml min-1 [1.73 m]-2, the urine albumin to creatinine ratio (uACR) was 6.9 mg/mmol, follow-up was 4.3 years and 21% had the composite endpoint. On cross-validation in derivation (n = 686), KidneyIntelX had an AUC of 0.77 (95% CI 0.74, 0.79). In validation (n = 460), the AUC was 0.77 (95% CI 0.76, 0.79). By comparison, the AUC for the clinical model was 0.62 (95% CI 0.61, 0.63) in derivation and 0.61 (95% CI 0.60, 0.63) in validation. Using derivation cut-offs, KidneyIntelX stratified 46%, 37% and 17% of the validation cohort into low-, intermediate- and high-risk groups for the composite kidney endpoint, respectively. The PPV for progressive decline in kidney function in the high-risk group was 61% for KidneyIntelX vs 40% for the highest risk strata by KDIGO categorisation (p < 0.001). Only 10% of those scored as low risk by KidneyIntelX experienced progression (i.e., NPV of 90%). The NRIevent for the high-risk group was 41% (p < 0.05).
Conclusions: KidneyIntelX improved prediction of kidney outcomes over KDIGO and clinical models in individuals with early stages of DKD.
Keywords: Biomarkers; Diabetic kidney disease; Electronic data; Machine learning; Prediction.
Figures
Fig. 1
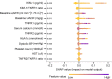
Shapley additive explanations (SHAP) plot showing relative feature importance. SHAP summary plots order features based on their importance. Each plot is made up of individual points from the training dataset with a higher value being darker purple and a lower value being more yellow. If the dots on one side of the middle line are more purple or yellow, this suggests that the values are increasing or decreasing, respectively, moving the prediction in that direction. For example, higher systolic BP is associated with higher risk of the composite kidney outcome. AST, aspartate aminotransferase
Fig. 2
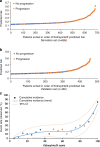
Composite kidney endpoint event rates by (a) KidneyIntelX predicted risk in derivation set, (b) KidneyIntelX predicted risk in validation set and (c) KidneyIntelX score prediction distributions of patients with DKD according to the risk of composite kidney endpoint in the derivation and validation set. (a, b) Events are denoted with an orange dot (progression) and represent the composite kidney endpoint within 5 years. Non-events are denoted with blue dots (no progression) and represent an absence of the composite kidney event in the follow-up period. (c) Dots represent cumulative incidence: blue, low risk 10% (6%, 14%); pink, intermediate risk 22% (16%, 28%); and red, high risk 61% (50%, 71%)
Fig. 3
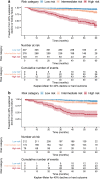
Kaplan–Meier curves by KidneyIntelX risk strata for the endpoint of sustained 40% decline in eGFR or kidney failure in derivation (a) and validation (b) sets. The risk cut-offs derived from derivation and applied to validation were: low risk 0–0.061129, intermediate risk 0.061129–0.30209 and high risk 0.30209–1. In the derivation set, 45% were low risk, 40% were intermediate risk and 15% were high risk. In the validation set, 46% were low risk, 37% were intermediate risk, and 17% were high risk. The HR for high vs low risk was 18.3 (95% CI 10.1, 33.1) in derivation and 14.7 (95% CI 7.8, 27.6) in validation. The HR for high vs intermediate risk was HR 5.7 (95% CI 3.7, 8.7) in derivation and 6.0 (95% CI 3.5, 10.0) in validation. The HR for high vs low and intermediate risk combined was 9.2 (95% CI 6.2, 13.6) in derivation and 9.1 (95% CI 5.8, 14.4) in validation
References
- USRDS (2018) Annual data report: atlas of chronic kidney disease and end-stage renal disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases
- KDIGO Clinical practice guideline for the evaluation and management of chronic kidney disease. Kidney Int Suppl. 2012;3:1–163.
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL085757/HL/NHLBI NIH HHS/United States
- U01 HG009610/HG/NHGRI NIH HHS/United States
- IK2 CX001780/CX/CSRD VA/United States
- R01 DK108803/DK/NIDDK NIH HHS/United States
- U01 OH011326/OH/NIOSH CDC HHS/United States
- R01 DK126477/DK/NIDDK NIH HHS/United States
- U01 DK106962/DK/NIDDK NIH HHS/United States
- R01 DK115562/DK/NIDDK NIH HHS/United States
- U01 HG007278/HG/NHGRI NIH HHS/United States
- R01 DK112258/DK/NIDDK NIH HHS/United States
- K23 DK107908/DK/NIDDK NIH HHS/United States
- U01 DK116100/DK/NIDDK NIH HHS/United States
- K23 DK124645/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous