Large-Scale Phylogenomic Analyses Reveal the Monophyly of Bryophytes and Neoproterozoic Origin of Land Plants - PubMed (original) (raw)
Large-Scale Phylogenomic Analyses Reveal the Monophyly of Bryophytes and Neoproterozoic Origin of Land Plants
Danyan Su et al. Mol Biol Evol. 2021.
Abstract
The relationships among the four major embryophyte lineages (mosses, liverworts, hornworts, vascular plants) and the timing of the origin of land plants are enigmatic problems in plant evolution. Here, we resolve the monophyly of bryophytes by improving taxon sampling of hornworts and eliminating the effect of synonymous substitutions. We then estimate the divergence time of crown embryophytes based on three fossil calibration strategies, and reveal that maximum calibration constraints have a major effect on estimating the time of origin of land plants. Moreover, comparison of priors and posteriors provides a guide for evaluating the optimal calibration strategy. By considering the reliability of fossil calibrations and the influences of molecular data, we estimate that land plants originated in the Precambrian (980-682 Ma), much older than widely recognized. Our study highlights the important contribution of molecular data when faced with contentious fossil evidence, and that fossil calibrations used in estimating the timescale of plant evolution require critical scrutiny.
Keywords: bryophytes; land plants; maximum bounds; phylogeny; timescale.
© The Author(s) 2021. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures
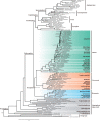
Fig. 1.
The concatenation species tree of land plants and their algal relatives. The cladogram is reconstructed using an amino acid supermatrix of 1,440 genes by IQ-TREE. Nodal support values are estimated by SH-aLRT test (SH) and ultrafast bootstrap (UFBS) in IQ-TREE, and Bayesian posterior probabilities (BPP) in ExaBayes. The first three are SH, UFBS, and BPP values based on codon-degenerate nucleotide data, and the last three are SH, UFBS, and BPP values based on amino acid data. The nodes without values indicate full support.
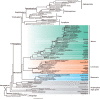
Fig. 2.
The coalescent species tree of land plants and their algal relatives. The cladogram is reconstructed using 1,440 genes by ASTRAL. Nodal support values are estimated by local posterior probability and multilocus bootstrapping (gene+site resampling) (PP/MLBS), and the nodes without values indicate full support.
Fig. 3.
Comparison of results from three different strategies. (a) Comparison of divergence time estimates from three fossil strategies. Node ages are plotted at the posterior means, with horizontal bars representing 95% credibility intervals. (b) Comparison of the age distributions on the crown node of land plants for the specified priors (black line), effective priors (purple), and posteriors (orange red).
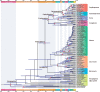
Fig. 4.
Timetree of Streptophyta inferred from Strategy 2. Node ages are plotted at the posterior means and horizontal bars represent 95% credibility intervals. A total of 22 fossil calibration nodes are marked by red dots.
Similar articles
- A review of molecular-clock calibrations and substitution rates in liverworts, mosses, and hornworts, and a timeframe for a taxonomically cleaned-up genus Nothoceros.
Villarreal JC, Renner SS. Villarreal JC, et al. Mol Phylogenet Evol. 2014 Sep;78:25-35. doi: 10.1016/j.ympev.2014.04.014. Epub 2014 Apr 30. Mol Phylogenet Evol. 2014. PMID: 24792087 - Organellomic data sets confirm a cryptic consensus on (unrooted) land-plant relationships and provide new insights into bryophyte molecular evolution.
Bell D, Lin Q, Gerelle WK, Joya S, Chang Y, Taylor ZN, Rothfels CJ, Larsson A, Villarreal JC, Li FW, Pokorny L, Szövényi P, Crandall-Stotler B, DeGironimo L, Floyd SK, Beerling DJ, Deyholos MK, von Konrat M, Ellis S, Shaw AJ, Chen T, Wong GK, Stevenson DW, Palmer JD, Graham SW. Bell D, et al. Am J Bot. 2020 Jan;107(1):91-115. doi: 10.1002/ajb2.1397. Epub 2019 Dec 8. Am J Bot. 2020. PMID: 31814117 - The hornwort genome and early land plant evolution.
Zhang J, Fu XX, Li RQ, Zhao X, Liu Y, Li MH, Zwaenepoel A, Ma H, Goffinet B, Guan YL, Xue JY, Liao YY, Wang QF, Wang QH, Wang JY, Zhang GQ, Wang ZW, Jia Y, Wang MZ, Dong SS, Yang JF, Jiao YN, Guo YL, Kong HZ, Lu AM, Yang HM, Zhang SZ, Van de Peer Y, Liu ZJ, Chen ZD. Zhang J, et al. Nat Plants. 2020 Feb;6(2):107-118. doi: 10.1038/s41477-019-0588-4. Epub 2020 Feb 10. Nat Plants. 2020. PMID: 32042158 Free PMC article. - The hornworts: morphology, evolution and development.
Frangedakis E, Shimamura M, Villarreal JC, Li FW, Tomaselli M, Waller M, Sakakibara K, Renzaglia KS, Szövényi P. Frangedakis E, et al. New Phytol. 2021 Jan;229(2):735-754. doi: 10.1111/nph.16874. Epub 2020 Sep 15. New Phytol. 2021. PMID: 32790880 Free PMC article. Review. - Major transitions in the evolution of early land plants: a bryological perspective.
Ligrone R, Duckett JG, Renzaglia KS. Ligrone R, et al. Ann Bot. 2012 Apr;109(5):851-71. doi: 10.1093/aob/mcs017. Epub 2012 Feb 22. Ann Bot. 2012. PMID: 22356739 Free PMC article. Review.
Cited by
- Phylotranscriptomics of liverworts: revisiting the backbone phylogeny and ancestral gene duplications.
Dong S, Yu J, Zhang L, Goffinet B, Liu Y. Dong S, et al. Ann Bot. 2022 Dec 31;130(7):951-964. doi: 10.1093/aob/mcac113. Ann Bot. 2022. PMID: 36075207 Free PMC article. - Insights into the conservation and diversification of the molecular functions of YTHDF proteins.
Flores-Téllez D, Tankmar MD, von Bülow S, Chen J, Lindorff-Larsen K, Brodersen P, Arribas-Hernández L. Flores-Téllez D, et al. PLoS Genet. 2023 Oct 10;19(10):e1010980. doi: 10.1371/journal.pgen.1010980. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37816028 Free PMC article. - Plant Kinesin Repertoires Expand with New Domain Architecture and Contract with the Loss of Flagella.
Lucas J, Geisler M. Lucas J, et al. J Mol Evol. 2024 Aug;92(4):381-401. doi: 10.1007/s00239-024-10178-9. Epub 2024 Jun 26. J Mol Evol. 2024. PMID: 38926179 - Plant protein-coding gene families: Their origin and evolution.
Fang Y, Jiang J, Hou X, Guo J, Li X, Zhao D, Xie X. Fang Y, et al. Front Plant Sci. 2022 Sep 7;13:995746. doi: 10.3389/fpls.2022.995746. eCollection 2022. Front Plant Sci. 2022. PMID: 36160967 Free PMC article. Review. - Genome-wide identification and evolutionary view of ALOG gene family in Solanaceae.
Turchetto C, Silvério AC, Waschburger EL, Lacerda MEG, Quintana IV, Turchetto-Zolet AC. Turchetto C, et al. Genet Mol Biol. 2023 Nov 24;46(3 Suppl 1):e20230142. doi: 10.1590/1415-4757-GMB-2023-0142. eCollection 2023. Genet Mol Biol. 2023. PMID: 38048778 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials