No Evidence for Human Monocyte-Derived Macrophage Infection and Antibody-Mediated Enhancement of SARS-CoV-2 Infection - PubMed (original) (raw)
No Evidence for Human Monocyte-Derived Macrophage Infection and Antibody-Mediated Enhancement of SARS-CoV-2 Infection
Obdulio García-Nicolás et al. Front Cell Infect Microbiol. 2021.
Abstract
Vaccines are essential to control the spread of severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) and to protect the vulnerable population. However, one safety concern of vaccination is the possible development of antibody-dependent enhancement (ADE) of SARS-CoV-2 infection. The potential infection of Fc receptor bearing cells such as macrophages, would support continued virus replication and inflammatory responses, and thereby potentially worsen the clinical outcome of COVID-19. Here we demonstrate that SARS-CoV-2 and SARS-CoV neither infect human monocyte-derived macrophages (hMDM) nor induce inflammatory cytokines in these cells, in sharp contrast to Middle East respiratory syndrome (MERS) coronavirus and the common cold human coronavirus 229E. Furthermore, serum from convalescent COVID-19 patients neither induced enhancement of SARS-CoV-2 infection nor innate immune response in hMDM. Although, hMDM expressed angiotensin-converting enzyme 2, no or very low levels of transmembrane protease serine 2 were found. These results support the view that ADE may not be involved in the immunopathological processes associated with COVID-19, however, more studies are necessary to understand the potential contribution of antibodies-virus complexes with other cells expressing FcR receptors.
Keywords: ADE; COVID-19 convalescent sera; SARS-CoV-2; human coronaviruses; human monocyte-derived macrophages.
Copyright © 2021 García-Nicolás, V’kovski, Zettl, Zimmer, Thiel and Summerfield.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
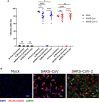
Figure 1
Determination of SARS-CoV and SARS-CoV-2 infected hMDM by immunolabeling for dsRNA and N protein. Vere E6 cells were infected with SARS-CoV and SARS-CoV-2 at MOI 1 TCID50/cell, and after 24 hpi dsRNA and N protein were labeled with specific antibodies. The nuclei were stained with DAPI. Then positive cells for dsRNA and N were quantified either by flow cytometry or immunofluorescence microscopy (A). In (B) example of representative images acquired by fluorescence microscopy is shown. The scale bar represent 40 µm. The data are from three independent experiments. Statistically significant differences between the conditions are indicated by asterisks (ns indicates non-statistical differences, *p < 0.05, **p ≤ 0.002 and ****p ≤ 0.0001).
Figure 2
Susceptibility of hMDM to different human coronaviruses. Human MDM were inoculated with different coronaviruses (hCoV-229E, MERS-CoV, SARS-CoV-1 and SARS-CoV-2) using an MOI of 1 TCID50/cell. Mock-infected cells were included as controls. After incubating the cells for 1.5 hours, the inoculum was removed, the cells washed, and fresh medium added. At 24 hpi, dsRNA in the cells was detected with a specific antibody and nuclei were stained with DAPI; the scale bar represents 40 µm. (A) The percentage of dsRNA-positive hMDM was calculated for 10 fields per condition (B). In (C) virus titers are shown. The same experiment was repeated with hCoV-229E, SARS-CoV-1 and SARS-CoV-2, and infected cells were quantified at 24, 48 and 72 hpi (D). The relative number of total hMDM per well was calculated taking as reference the number of cells at 24 hpi (E). The data from three independent experiments run in triplicates are shown in each panel. Statistically significant differences between the conditions are indicated by different superscript letters in (B, C) (p < 0.05), and by asterisks in (D, E) (****p ≤ 0.0001).
Figure 3
Human MDM immune response after coronavirus infection. Human MDM were inoculated with different coronaviruses (hCoV-229E, MERS-CoV, SARS-CoV-1 and SARS-CoV-2) as described before. Mock-infected cells or cells treated with LPS or poly I:C served as controls. After 24 hpi TNF (A) IFN-β (B) and IL-6 (C) were determined in the cell culture supernatants. The data from three independent experiments run in triplicates are shown. Different superscript letters indicate a significant difference (p < 0.05) between the conditions.
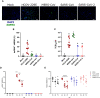
Figure 4
ACE2 and TMPRSS2 expression in hMDM. After 6 days of differentiation ACE2 and TMPRSS2 were immunolabeled with specific antibodies and positive cells were assessed by flow cytometry. A549 cells transfected with ACE2 and TMPRSS2 were used as control (A). Representative histograms for each marker in the analyzed cells are shown (B). The data from 5 different human donor hMDM run in triplicates are shown. Statistically significant differences in the expression of each marker between both cell types are marked by asterisks (****p ≤ 0.0001).
Similar articles
- Two Different Antibody-Dependent Enhancement (ADE) Risks for SARS-CoV-2 Antibodies.
Ricke DO. Ricke DO. Front Immunol. 2021 Feb 24;12:640093. doi: 10.3389/fimmu.2021.640093. eCollection 2021. Front Immunol. 2021. PMID: 33717193 Free PMC article. - Antibody-Dependent Enhancement of SARS-CoV-2 Infection Is Mediated by the IgG Receptors FcγRIIA and FcγRIIIA but Does Not Contribute to Aberrant Cytokine Production by Macrophages.
Maemura T, Kuroda M, Armbrust T, Yamayoshi S, Halfmann PJ, Kawaoka Y. Maemura T, et al. mBio. 2021 Oct 26;12(5):e0198721. doi: 10.1128/mBio.01987-21. Epub 2021 Sep 28. mBio. 2021. PMID: 34579572 Free PMC article. - Multiple Routes of Antibody-Dependent Enhancement of SARS-CoV-2 Infection.
Okuya K, Hattori T, Saito T, Takadate Y, Sasaki M, Furuyama W, Marzi A, Ohiro Y, Konno S, Hattori T, Takada A. Okuya K, et al. Microbiol Spectr. 2022 Apr 27;10(2):e0155321. doi: 10.1128/spectrum.01553-21. Epub 2022 Mar 23. Microbiol Spectr. 2022. PMID: 35319248 Free PMC article. - Human and novel coronavirus infections in children: a review.
Rajapakse N, Dixit D. Rajapakse N, et al. Paediatr Int Child Health. 2021 Feb;41(1):36-55. doi: 10.1080/20469047.2020.1781356. Epub 2020 Jun 25. Paediatr Int Child Health. 2021. PMID: 32584199 Review. - Antibody-dependent enhancement of coronavirus.
Wen J, Cheng Y, Ling R, Dai Y, Huang B, Huang W, Zhang S, Jiang Y. Wen J, et al. Int J Infect Dis. 2020 Nov;100:483-489. doi: 10.1016/j.ijid.2020.09.015. Epub 2020 Sep 11. Int J Infect Dis. 2020. PMID: 32920233 Free PMC article. Review.
Cited by
- CD169-mediated restrictive SARS-CoV-2 infection of macrophages induces pro-inflammatory responses.
Jalloh S, Olejnik J, Berrigan J, Nisa A, Suder EL, Akiyama H, Lei M, Ramaswamy S, Tyagi S, Bushkin Y, Mühlberger E, Gummuluru S. Jalloh S, et al. PLoS Pathog. 2022 Oct 24;18(10):e1010479. doi: 10.1371/journal.ppat.1010479. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36279285 Free PMC article. - A rapid real-time polymerase chain reaction-based live virus microneutralization assay for detection of neutralizing antibodies against SARS-CoV-2 in blood/serum.
Abidi SH, Imtiaz K, Kanji A, Qaiser S, Khan E, Iqbal K, Veldhoen M, Ghias K, Simas JP, Hasan Z. Abidi SH, et al. PLoS One. 2021 Dec 10;16(12):e0259551. doi: 10.1371/journal.pone.0259551. eCollection 2021. PLoS One. 2021. PMID: 34890401 Free PMC article. - Interactions of adenoviruses with platelets and coagulation and the vaccine-induced immune thrombotic thrombocytopenia syndrome.
Gresele P, Momi S, Marcucci R, Ramundo F, De Stefano V, Tripodi A. Gresele P, et al. Haematologica. 2021 Dec 1;106(12):3034-3045. doi: 10.3324/haematol.2021.279289. Haematologica. 2021. PMID: 34407607 Free PMC article. Review. - Prior COVID-19 Immunization Does Not Cause IgA- or IgG-Dependent Enhancement of SARS-CoV-2 Infection.
Yaugel-Novoa M, Noailly B, Jospin F, Berger AE, Waeckel L, Botelho-Nevers E, Longet S, Bourlet T, Paul S. Yaugel-Novoa M, et al. Vaccines (Basel). 2023 Mar 31;11(4):773. doi: 10.3390/vaccines11040773. Vaccines (Basel). 2023. PMID: 37112685 Free PMC article. - Have Diagnostics, Therapies, and Vaccines Made the Difference in the Pandemic Evolution of COVID-19 in Comparison with "Spanish Flu"?
Lista F, Peragallo MS, Biselli R, De Santis R, Mariotti S, Nisini R, D'Amelio R. Lista F, et al. Pathogens. 2023 Jun 23;12(7):868. doi: 10.3390/pathogens12070868. Pathogens. 2023. PMID: 37513715 Free PMC article. Review.
References
- Dijkman R., Jebbink M. F., Koekkoek S. M., Deijs M., Jonsdottir H. R., Molenkamp R., et al. . (2013). Isolation and characterization of current human coronavirus strains in primary human epithelial cell cultures reveal differences in target cell tropism. J. Virol. 87 (11), 6081–6090. 10.1128/JVI.03368-12 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous