A global resource for genomic predictions of antimicrobial resistance and surveillance of Salmonella Typhi at pathogenwatch - PubMed (original) (raw)
. 2021 May 17;12(1):2879.
doi: 10.1038/s41467-021-23091-2.
Corin A Yeats 2, Richard J Goater 3 4, Khalil Abudahab 3, Benjamin Taylor 2, Anthony Underwood 3, Leonor Sánchez-Busó 2 5, Vanessa K Wong 6, Zoe A Dyson 6 7 8, Satheesh Nair 9, Se Eun Park 10, Florian Marks 10, Andrew J Page 11 12, Jacqueline A Keane 11, Stephen Baker 13, Kathryn E Holt 7 8, Gordon Dougan 6, David M Aanensen 14 15
Affiliations
- PMID: 34001879
- PMCID: PMC8128892
- DOI: 10.1038/s41467-021-23091-2
A global resource for genomic predictions of antimicrobial resistance and surveillance of Salmonella Typhi at pathogenwatch
Silvia Argimón et al. Nat Commun. 2021.
Abstract
As whole-genome sequencing capacity becomes increasingly decentralized, there is a growing opportunity for collaboration and the sharing of surveillance data within and between countries to inform typhoid control policies. This vision requires free, community-driven tools that facilitate access to genomic data for public health on a global scale. Here we present the Pathogenwatch scheme for Salmonella enterica serovar Typhi (S. Typhi), a web application enabling the rapid identification of genomic markers of antimicrobial resistance (AMR) and contextualization with public genomic data. We show that the clustering of S. Typhi genomes in Pathogenwatch is comparable to established bioinformatics methods, and that genomic predictions of AMR are highly concordant with phenotypic susceptibility data. We demonstrate the public health utility of Pathogenwatch with examples selected from >4,300 public genomes available in the application. Pathogenwatch provides an intuitive entry point to monitor of the emergence and spread of S. Typhi high risk clones.
Conflict of interest statement
The authors declare no competing interests.
Figures
Fig. 1. Workflow of the Typhi Pathogenwatch application.
Input assemblies or sequence reads and metadata files can be uploaded via drag-and-drop onto the Upload page. Once the analyses completed, the genomes are listed on the Genomes page with Pathogenwatch outputs for speciation and MLST. Clicking on a genome name on the list pops up a Genome Report. The user can create collections of genomes. The Collection view displays the user genomes clustered by genetic similarity on a tree, their location on a map, a timeline, as well as tables for metadata, typing and AMR. The Population view displays the user genomes by their closest reference genome in the population tree. Clicking on one of the highlighted nodes (purple triangles) opens the Population subtree view, which contextualizes the user genomes with the closest public genomes.
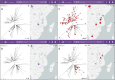
Fig. 2. Pathogenwatch provides genomic context for outbreak investigations.
a, b Genomes from an outbreak in Zambia (purple markers on tree and map) are linked by genetic relatedness to genomes from neighboring countries Malawi and Tanzania (gray markers) forming two separate groups containing 16 (a) and 4 (b) outbreak genomes, respectively. The number of pairwise differences (range) between outbreak and related genomes in the Pathogenwatch score matrix are indicated on the bottom-right of the tree panel. c, d Differential distribution of trimethoprim resistance genes dfrA7 (c) and dfrA14 (d) across the two clades containing outbreak genomes. The presence of the dfr genes is indicated in red on the tree and map. The data are available at
https://pathogen.watch/collection/g5pbucot6e58-hendriksen-et-al-2015
.
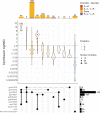
Fig. 3. Genotypic predictions of antimicrobial resistance.
Distribution of minimum inhibitory concentration (MIC) values (mg L−1) for ciprofloxacin in a collection of S. Typhi isolates with different combinations of genetic mechanisms that are known to confer resistance to this antibiotic. Only combinations observed in at least five genomes are shown. Dashed horizontal lines on the violin plots mark the CLSI clinical breakpoint for ciprofloxacin. Point colors inside violins represent the genotypic AMR prediction by Pathogenwatch on each combination of mechanisms. Barplots on the top show the abundance of genomes with each combination of mechanisms. Bar colors represent the differences between the predicted and the observed SIR (e.g., red for a predicted susceptible mechanism when the observed phenotype is resistant). S susceptible, I intermediate, and R resistant.
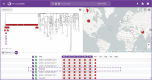
Fig. 4. Pathogenwatch data reusability.
Fifteen genomes carrying the acrB_R717Q mutation recently linked to azithromycin resistance in S. Typhi are shown in red on the tree of 4389 public genomes and on the map. The presence of the mutation is also indicated by the red circles on the SNPs table. Three of these genomes (tree labels) belong to isolates collected before the mutation was first described and are shown in more detail in the bottom panels. The data are available at
https://pathogen.watch/collection/07lsscrbhu2x-public-genomes
.
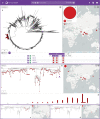
Fig. 5. Rapid risk assessment of typhoid fever cases in non-endemic regions.
Pathogenwatch places genome PHL5950 from an isolate recovered in Canada and with travel history to Pakistan within the XDR-outbreak in Pakistan. Red markers on the tree and table indicate XDR isolates. The data are available at
https://pathogen.watch/collection/11lsok8nrzts-wong-et-al-2018-idcases-15e00492
.
Fig. 6. Pathogenwatch to for collaborative international surveillance of S. Typhi.
a Pathogenwatch highlights 195 ciprofloxacin-resistant triple mutants on the public data tree and map by simultaneously selecting the mutations gyrA_S83F, gyrA_D87N, and parC_S80I on the SNPs table (red markers). b Detailed visualization of the triple mutants showing the temporal distribution of the genomes on the timeline. Purple arrowhead: four genomes with sul1, dfrA15, tetA(A) and the IncN replicon from the UK and Japan. Selecting individual clades on the tree shows distinct clades that span neighboring countries India-Pakistan (c) and India-Nepal (d). The data are available at
https://pathogen.watch/collection/07lsscrbhu2x-public-genomes
.
Similar articles
- Salmonella enterica Serovar Typhi in Bangladesh: Exploration of Genomic Diversity and Antimicrobial Resistance.
Tanmoy AM, Westeel E, De Bruyne K, Goris J, Rajoharison A, Sajib MSI, van Belkum A, Saha SK, Komurian-Pradel F, Endtz HP. Tanmoy AM, et al. mBio. 2018 Nov 13;9(6):e02112-18. doi: 10.1128/mBio.02112-18. mBio. 2018. PMID: 30425150 Free PMC article. - Salmonella Typhi genotypic diversity, cluster identification and antimicrobial resistance determinants in Mukuru settlement, Nairobi Kenya.
Kasiano P, Morita M, Kodama T, Hiyoshi H, Kavai S, Kiiru S, Kariuki S. Kasiano P, et al. BMC Infect Dis. 2024 Jul 24;24(1):727. doi: 10.1186/s12879-024-09635-z. BMC Infect Dis. 2024. PMID: 39048963 Free PMC article. - A Race against Time: Reduced Azithromycin Susceptibility in Salmonella enterica Serovar Typhi in Pakistan.
Iqbal J, Dehraj IF, Carey ME, Dyson ZA, Garrett D, Seidman JC, Kabir F, Saha S, Baker S, Qamar FN. Iqbal J, et al. mSphere. 2020 Jul 22;5(4):e00215-20. doi: 10.1128/mSphere.00215-20. mSphere. 2020. PMID: 32699118 Free PMC article. - Antimicrobial resistance in Salmonella enterica serovar typhi and paratyphi in South Asia-current status, issues and prospects.
Akhtar S, Sarker MR, Jabeen K, Sattar A, Qamar A, Fasih N. Akhtar S, et al. Crit Rev Microbiol. 2015;41(4):536-45. doi: 10.3109/1040841X.2014.880662. Epub 2014 Mar 19. Crit Rev Microbiol. 2015. PMID: 24645636 Review. - Salmonella Typhi genomics: envisaging the future of typhoid eradication.
Yap KP, Thong KL. Yap KP, et al. Trop Med Int Health. 2017 Aug;22(8):918-925. doi: 10.1111/tmi.12899. Epub 2017 Jun 19. Trop Med Int Health. 2017. PMID: 28544285 Review.
Cited by
- Genomic characterization of invasive typhoidal and non-typhoidal Salmonella in southwestern Nigeria.
Ikhimiukor OO, Oaikhena AO, Afolayan AO, Fadeyi A, Kehinde A, Ogunleye VO, Aboderin AO, Oduyebo OO, Elikwu CJ, Odih EE, Komolafe I, Argimón S, Egwuenu A, Adebiyi I, Sadare OA, Okwor T, Kekre M, Underwood A, Ihekweazu C, Aanensen DM, Okeke IN. Ikhimiukor OO, et al. PLoS Negl Trop Dis. 2022 Aug 26;16(8):e0010716. doi: 10.1371/journal.pntd.0010716. eCollection 2022 Aug. PLoS Negl Trop Dis. 2022. PMID: 36026470 Free PMC article. - Persistence of Rare Salmonella Typhi Genotypes Susceptible to First-Line Antibiotics in the Remote Islands of Samoa.
Sikorski MJ, Hazen TH, Desai SN, Nimarota-Brown S, Tupua S, Sialeipata M, Rambocus S, Ingle DJ, Duchene S, Ballard SA, Valcanis M, Zufan S, Ma J, Sahl JW, Maes M, Dougan G, Thomsen RE, Robins-Browne RM, Howden BP, Naseri TK, Levine MM, Rasko DA. Sikorski MJ, et al. mBio. 2022 Oct 26;13(5):e0192022. doi: 10.1128/mbio.01920-22. Epub 2022 Sep 12. mBio. 2022. PMID: 36094088 Free PMC article. - Occurrence of Multidrug-Resistant Bacteria Resulting from the Selective Pressure of Antibiotics: A Comprehensive Analysis of ESBL K. pneumoniae and MRSP Isolated in a Dog with Rhinorrhea.
Rodrigues IC, Ribeiro-Almeida M, Ribeiro J, Silveira L, Prata JC, Pista A, Martins da Costa P. Rodrigues IC, et al. Vet Sci. 2023 May 2;10(5):326. doi: 10.3390/vetsci10050326. Vet Sci. 2023. PMID: 37235409 Free PMC article. - Genome-Based Assessment of Antimicrobial Resistance and Virulence Potential of Isolates of Non-Pullorum/Gallinarum Salmonella Serovars Recovered from Dead Poultry in China.
Li Y, Kang X, Ed-Dra A, Zhou X, Jia C, Müller A, Liu Y, Kehrenberg C, Yue M. Li Y, et al. Microbiol Spectr. 2022 Aug 31;10(4):e0096522. doi: 10.1128/spectrum.00965-22. Epub 2022 Jun 21. Microbiol Spectr. 2022. PMID: 35727054 Free PMC article. - Genomic Characterization of Salmonella Typhimurium Isolated from Guinea Pigs with Salmonellosis in Lima, Peru.
Carhuaricra Huaman DE, Luna Espinoza LR, Rodríguez Cueva CL, Duran Gonzales CG, Rosadio Alcántara RH, Setubal JC, Maturrano Hernández L. Carhuaricra Huaman DE, et al. Microorganisms. 2022 Aug 27;10(9):1726. doi: 10.3390/microorganisms10091726. Microorganisms. 2022. PMID: 36144328 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials