Why is primary endosymbiosis so rare? - PubMed (original) (raw)
Review
. 2021 Sep;231(5):1693-1699.
doi: 10.1111/nph.17478. Epub 2021 Jun 21.
Affiliations
- PMID: 34018613
- PMCID: PMC8711089
- DOI: 10.1111/nph.17478
Review
Why is primary endosymbiosis so rare?
Timothy G Stephens et al. New Phytol. 2021 Sep.
Abstract
Endosymbiosis is a relationship between two organisms wherein one cell resides inside the other. This affiliation, when stable and beneficial for the 'host' cell, can result in massive genetic innovation with the foremost examples being the evolution of eukaryotic organelles, the mitochondria and plastids. Despite its critical evolutionary role, there is limited knowledge about how endosymbiosis is initially established and how host-endosymbiont biology is integrated. Here, we explore this issue, using as our model the rhizarian amoeba Paulinella, which represents an independent case of primary plastid origin that occurred c. 120 million yr ago. We propose the 'chassis and engine' model that provides a theoretical framework for understanding primary plastid endosymbiosis, potentially explaining why it is so rare.
Keywords: Rhizaria; endosymbiotic gene transfer; genome reduction; organellogenesis; photosynthetic eukaryotes; primary endosymbiosis.
© 2021 The Authors. New Phytologist © 2021 New Phytologist Foundation.
Figures
Fig. 1
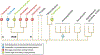
The history of plastid endosymbiosis in eukaryotes. This is a highly reduced tree that only shows the groups of interest (see also Ponce-Toledo et al., 2019). Primary plastid origin occurred in the ancestors of Archaeplastida and photosynthetic Paulinella (within Rhizaria). Green algal secondary endosymbiosis occurred independently in the chlorarachniophyte (Rhizaria) and Euglenozoa lineages. Red algal secondary endosymbiosis has occurred in the alveolates (e.g. dinoflagellates), stramenopiles, cryptophytes and haptophytes. These taxa are informally referred to as CRASH (i.e. cryptophytes, rhizarians, alveolates, stramenopiles and haptophytes) with phylogenetic evidence existing for the monophyly of SAR (stramenopiles, alveolates and rhizarians) taxa (Bhattacharya & Price, 2020; Fan et al., 2020). Broken lines indicate unclear phylogenetic affiliations that may impact the number of red algal endosymbiosis if some (or all, highly unlikely) of these lineages share a single event. There is a variety of plastid-lacking heterotrophic lineages, not shown here, such as ciliates, telonemids and katablepharids, that are sister to photosynthetic taxa in the CRASH. Multiple tertiary endosymbioses involving green algae, haptophytes and diatoms have occurred in the dinoflagellates that are not shown here (Gross et al., 2012).
Fig. 2
Putative timeline of plastid evolution in Paulinella encompassing both a historical and future perspective based on the chassis and engine model. (a) Heterotrophic Paulinella ingesting cyanobacterial cells likely using filose, feeding pseudopodia as a result of the presence of silica scales on the cell surface (Johnson et al., 1988). DNA from ingested bacterial cells can enter the nucleus and become integrated into the host genome via horizontal gene transfer (HGT). These foreign genes, as well as other novel ‘dark’ genes that evolved in this lineage, enabled the host to maintain the cyanobacteria in food vacuoles for increasingly longer periods of time until the association became permanent. (b) The status of extant photosynthetic Paulinella showing the two chromatophores and current evolutionary processes. (c) Putative future status of photosynthetic Paulinella. The chromatophore will encode only essential genes that cannot be transferred to the host genome because the encoded proteins either are problematic to transport into the chromatophore or require redox regulationwithin the organelle(e.g. co-locationfor redox regulation (CoRR hypothesis); Allen, 2017).The chromatophore may also lose its peptidoglycan layer, as observed in other lineages containing primary plastids (except Glaucophyta). Adaptations will probably increase the Paulinella growth rate and the ability to deal with high light, expanding its environmental niche. (d) The chassis and engine model of plastid endosymbiosis. The approximate size of the chromatophore genome and the number of genes it encodes across Paulinella species are shown inside the engine. Parts of the model where questions remain about the endosymbiosis in Paulinella are shown in red text (for more detail see Box 1). crTP, chromatophore targeting peptide; EGT, endosymbiotic gene transfer; mt, mitochondrion; ROS, reactive oxygen species.
Similar articles
- Retrotransposition facilitated the establishment of a primary plastid in the thecate amoeba Paulinella.
Calatrava V, Stephens TG, Gabr A, Bhaya D, Bhattacharya D, Grossman AR. Calatrava V, et al. Proc Natl Acad Sci U S A. 2022 Jun 7;119(23):e2121241119. doi: 10.1073/pnas.2121241119. Epub 2022 May 31. Proc Natl Acad Sci U S A. 2022. PMID: 35639693 Free PMC article. - Paulinella, a model for understanding plastid primary endosymbiosis.
Gabr A, Grossman AR, Bhattacharya D. Gabr A, et al. J Phycol. 2020 Aug;56(4):837-843. doi: 10.1111/jpy.13003. Epub 2020 May 5. J Phycol. 2020. PMID: 32289879 Free PMC article. Review. - Endosymbiotic ratchet accelerates divergence after organelle origin: The Paulinella model for plastid evolution: The Paulinella model for plastid evolution.
Bhattacharya D, Etten JV, Benites LF, Stephens TG. Bhattacharya D, et al. Bioessays. 2023 Jan;45(1):e2200165. doi: 10.1002/bies.202200165. Epub 2022 Nov 3. Bioessays. 2023. PMID: 36328783 - Genomics-Informed Insights into Endosymbiotic Organelle Evolution in Photosynthetic Eukaryotes.
Nowack ECM, Weber APM. Nowack ECM, et al. Annu Rev Plant Biol. 2018 Apr 29;69:51-84. doi: 10.1146/annurev-arplant-042817-040209. Epub 2018 Feb 28. Annu Rev Plant Biol. 2018. PMID: 29489396 Review. - The Photosynthetic Adventure of Paulinella Spp.
Gagat P, Sidorczuk K, Pietluch F, Mackiewicz P. Gagat P, et al. Results Probl Cell Differ. 2020;69:353-386. doi: 10.1007/978-3-030-51849-3_13. Results Probl Cell Differ. 2020. PMID: 33263879
Cited by
- Loss of key endosymbiont genes may facilitate early host control of the chromatophore in Paulinella.
Gabr A, Stephens TG, Bhattacharya D. Gabr A, et al. iScience. 2022 Aug 17;25(9):104974. doi: 10.1016/j.isci.2022.104974. eCollection 2022 Sep 16. iScience. 2022. PMID: 36093053 Free PMC article. - Repeated horizontal acquisition of lagriamide-producing symbionts in Lagriinae beetles.
Uppal S, Waterworth SC, Nick A, Vogel H, Flórez LV, Kaltenpoth M, Kwan JC. Uppal S, et al. bioRxiv [Preprint]. 2024 Jul 9:2024.01.23.576914. doi: 10.1101/2024.01.23.576914. bioRxiv. 2024. PMID: 39026795 Free PMC article. Updated. Preprint. - Costs of photosynthesis and cellular remodeling in trophic transitions of the unicellular red alga Galdieria partita.
Yamashita S, Hirooka S, Fujiwara T, Zhou B, Yagisawa F, Tamashiro K, Murakami H, Awai K, Miyagishima SY. Yamashita S, et al. Commun Biol. 2025 Jun 7;8(1):891. doi: 10.1038/s42003-025-08284-5. Commun Biol. 2025. PMID: 40483364 Free PMC article. - Organellar Evolution: A Path from Benefit to Dependence.
Oborník M. Oborník M. Microorganisms. 2022 Jan 7;10(1):122. doi: 10.3390/microorganisms10010122. Microorganisms. 2022. PMID: 35056571 Free PMC article. Review. - Evidence of a putative CO2 delivery system to the chromatophore in the photosynthetic amoeba Paulinella.
Gabr A, Stephens TG, Reinfelder JR, Liau P, Calatrava V, Grossman AR, Bhattacharya D. Gabr A, et al. Environ Microbiol Rep. 2024 Jun;16(3):e13304. doi: 10.1111/1758-2229.13304. Environ Microbiol Rep. 2024. PMID: 38923306 Free PMC article.
References
- Allen JF. 2017. The CoRR hypothesis for genes in organelles. Journal of Theoretical 434: 50–57. - PubMed
- Arias-Agudelo LM, Gonzalez F, Isaza JP, Alzate JF, Pabon-Mora N. 2019. Plastome reduction and gene content in New World Pilostyles (Apodanthaceae) unveils high similarities to African and Australian congeners. Molecular Phylogenetics and Evolution 135: 193–202. - PubMed
- Bhattacharya D, Price DC. 2020. The algal tree of life from a genomics perspective.In: Larkum AWD, Grossman AR, Raven JA, eds. Photosynthesis in algae: biochemical and physiological mechanisms. Cham, Switzerland: Springer International, 11–24.
- Bhaya D, Dufresne A, Vaulot D, Grossman A. 2002. Analysis of the hli gene family in marine and freshwater cyanobacteria. FEMS Microbiology Letters 215: 209–219. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources