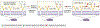Dismantling the bacterial glycocalyx: Chemical tools to probe, perturb, and image bacterial glycans - PubMed (original) (raw)
Review
Dismantling the bacterial glycocalyx: Chemical tools to probe, perturb, and image bacterial glycans
Phuong Luong et al. Bioorg Med Chem. 2021.
Abstract
The bacterial glycocalyx is a quintessential drug target comprised of structurally distinct glycans. Bacterial glycans bear unusual monosaccharide building blocks whose proper construction is critical for bacterial fitness, survival, and colonization in the human host. Despite their appeal as therapeutic targets, bacterial glycans are difficult to study due to the presence of rare bacterial monosaccharides that are linked and modified in atypical manners. Their structural complexity ultimately hampers their analytical characterization. This review highlights recent advances in bacterial chemical glycobiology and focuses on the development of chemical tools to probe, perturb, and image bacterial glycans and their biosynthesis. Current technologies have enabled the study of bacterial glycosylation machinery even in the absence of detailed structural information.
Keywords: Bacteria; Bioorthogonal chemistry; Chemical tools; Glycans; Inhibit; Lectin.
Copyright © 2021 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
Figure 1.. Structures of common bacterial glycans.
Exclusively bacterial monosaccharides are highlighted in color, with known species distribution in parentheses. Abbreviations: heptose = L-glycero-
d
-mannoheptose; KDO = 3-deoxy-
d
-manno-oct-2-ulosonic acid; AAT = 2-acetamido-4-amino-2,4,6-trideoxyhexose; gal_f_ = galactofuranose; MurNAc = muramic acid; Pse = pseudaminic acid; FucNAc = _N_-acetylfucosamine; Leg = legionaminic acid; Bac = bacillosamine.
Figure 2.. General schematic of metabolic oligosaccharide engineering (MOE).
First, a sugar modified with a chemical handle is metabolically incorporated via permissive glycosyltransferases (GTs) into cellular glycans. Second, an exquisitely selective partner undergoes bioorthogonal reaction with the chemical handle to yield a detectable covalent adduct bearing a reporter.
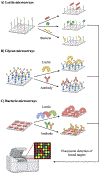
Figure 3.. Metabolic labeling of bacterial glycans with a variety of unnatural substrates.
Shown here is a panel of substrate analogs accessed from A) rare and common sugar building blocks, B) D-amino acids, and C) glycolipids. For each scaffold, chemical modification is permissible at positions with R groups and known metabolic fate indicated in parentheses. Abbreviations: MurNAc = muramic acid; KDO = 3-deoxy-
d
-manno-oct-2-ulosonic acid; AltdiNAc = 2,4-diacetamido-2,4,6-trideoxy-
l
-altrose; D-FucNAc = _N_-acetyl-
d
-fucosamine; DATDG = 2,4-diacetamido-2,4,6-trideoxyhexose; Bac-diNAc = N,_N’_-diacetylbacillosamine; GalNAc = _N-_acetylgalactosamine; GlcNAc = _N_-acetylglucosamine; AlkTMM = alkylated trehalose monomycolate.
Figure 4.. Microarray assays probe glycan-binding events pertinent to host-pathogen interactions.
A) Lectin microarrays differentiate bacteria based on distinct binding patterns. B) Glycan microarrays elucidate glycan epitopes key to host adhesion and identify novel immunogenic targets. C) Bacteria microarrays reveal intact glycan-binding interactions on bacterial surfaces.
Figure 5.. Mechanisms of action of bacterial glycosylation inhibitors and representative structures of each class of inhibitors.
A) Small molecule inhibitors compete with natural substrates in the active site of the target glycosyltransferase (GT), leading to the reduction of glycan production in the associated glycan biosynthetic pathway. B) Structures of representative small molecule inhibitors accessed from thienopyrimidine-6-carboxylate core-based analogs.C) Phosphoglycosyl transferases (PGTs) initiate glycan biosynthesis on a lipid carrier by catalyzing the transfer of a nucleoside sugar to a polyprenyl phosphate. Uridine analogs inhibit PGT activity and block addition of monosaccharides onto the lipid carrier. D) Structures of representative nucleoside inhibitors accessed from uridine analogs.E) Metabolic inhibitors consist of two classes: fluorosugars (chain terminators) and benzyl glycosides (substrate decoys). Fluorosugars terminate glycan elongation, while benzyl glycosides mimic endogenous glycan acceptors and divert glycan biosynthesis onto decoy substrates. F) Structures of representative metabolic inhibitors accessed from rare deoxy amino sugars, featuring chain terminators and substrate decoys. Abbreviations: Bac = bacillosamine (blue hexagon); DAT = 2,4-diacetamido-2,4,6-trideoxygalactose; FucNAc = _N_-acetylfucosamine.
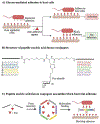
Figure 6.. Blocking glycan binding events key to pathogenesis.
A) Bacterial adhesion to host cells often depends on host-pathogen interactions. Anti-adhesive agents that bind to either bacterial adhesins or host receptors can block these glycan-mediated binding events, resulting in reduced infection. B) Structure of representative synthetic anti-adhesive compounds, featuring peptide nucleic acid-fucose conjugates. C) Peptide nucleic acid-fucose conjugates can form assemblies that bind to bacterial adhesins in a multivalent fashion and block bacterial entry to the host. Images B and C are inspired by Figure 2 from Machida et al.
Figure 7.. Approaches to potentiate antibiotics against resistant bacteria.
A) Inhibiting polysaccharide production during biofilm formation sensitizes bacteria to antibiotics by removing their protective barrier. B) Membrane-altering agents weaken the bacterial glycan coat and render bacteria vulnerable to antibiotics.
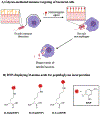
Figure 8.. Perturbing the bacterial glycocalyx to recruit immune responses.
A) Bacteria typically evade host immune detection by modifying surface glycans to block immune factors or mimic host antigens. Unnatural substrates bearing immune stimulants can exploit permissive metabolic pathways to become expressed on bacterial surfaces, ultimately recruiting macrophages to seek and destroy pathogens. B) Structures of representative immunogenic agents accessed from unnatural d-amino acids bearing 2.4-dinitrophenyl (DNP), which can be incorporated into the peptidoglycan and recruit anti-DNP antibodies.
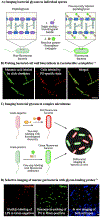
Figure 9.. Imaging strategies using bioorthogonal probes.
A) Sugar analogs bearing bioorthogonal groups illuminate bacterial glycans on target species. The sugars GlcNAc and MurNAc are depicted by blue squares and purple hexagons, respectively. B) Clickable muramic acid derivatives fluorescently labeled the peptidoglycan in Lactobacillus acidophilus. Reproduced with permission from DeMeester et al.C) Labeling bacteria via bioorthogonal probes alongside fluorescent antibiotics allows the differential imaging of Gram-negative and Gram-positive bacteria in vivo. D) Selective labeling of Gram-negative bacteria by 8-azido-8-deoxy-Kdo (8AzKdo, yellow hexagon) and Gram-positive bacteria by fluorescent vancomycin in the gut microbiome. Reproduced with permission from Wang et al.
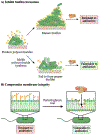
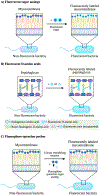
Figure 10.. Mechanisms of action of metabolic imaging probes.
A) Fluorescent sugar analogs are metabolically incorporated into endogenous glycans to enable their detection and tracking via fluorescence. Trehalose is depicted by two linked blue circles. B) Fluorescent and rotor-fluorogenic D-amino acids are metabolically incorporated into peptidoglycan and facilitate the real-time imaging of peptidoglycan biosynthesis. Rotor-fluorogenic D-amino acids turn fluorescent once incorporated into cellular peptidoglycan. C) Fluorophore-quencher probes accessed from sugar analogs become fluorescent upon specific enzyme-catalyzed reactions and allow the direct visualization of enzyme activity during cell wall biosynthesis.
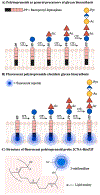
Figure 11.. Imaging bacterial glycans via fluorescent polyisoprenoids.
A) Bacterial polyisoprenoids act as lipid carriers upon which glycosyltransferases (GTs) construct higher order glycans, including polysaccharides. They are critical for biosynthesis of a number of glycans. B) A fluorescent reporter can be metabolically incorporated into bacterial polyisoprenoids to monitor glycan assembly steps and characterize glycosyltransferase activities. Glycans at different stages during biosynthesis can be imaged. C) Example of an imaging probe which consists of the fluorescent reporter 2-nitrileaniline appended to bactoprenyl phosphate. Figure adapted from Scott et al. Abbreviations: Ac = acetyl; Pyr = pyruvyl; 2CNA-B(nZ)P = 2-nitrileanilinobactoprenyl phosphate.
Similar articles
- Metabolic Glycan Labeling-Based Screen to Identify Bacterial Glycosylation Genes.
Moulton KD, Adewale AP, Carol HA, Mikami SA, Dube DH. Moulton KD, et al. ACS Infect Dis. 2020 Dec 11;6(12):3247-3259. doi: 10.1021/acsinfecdis.0c00612. Epub 2020 Nov 13. ACS Infect Dis. 2020. PMID: 33186014 Free PMC article. - Chemical biology tools to probe bacterial glycans.
Calles-Garcia D, Dube DH. Calles-Garcia D, et al. Curr Opin Chem Biol. 2024 Jun;80:102453. doi: 10.1016/j.cbpa.2024.102453. Epub 2024 Apr 5. Curr Opin Chem Biol. 2024. PMID: 38582017 Free PMC article. Review. - Metabolic inhibitors of bacterial glycan biosynthesis.
Williams DA, Pradhan K, Paul A, Olin IR, Tuck OT, Moulton KD, Kulkarni SS, Dube DH. Williams DA, et al. Chem Sci. 2020 Jan 8;11(7):1761-1774. doi: 10.1039/c9sc05955e. Chem Sci. 2020. PMID: 34123271 Free PMC article. - Nanoscale materials for probing the biological functions of the glycocalyx.
Huang ML, Godula K. Huang ML, et al. Glycobiology. 2016 Aug;26(8):797-803. doi: 10.1093/glycob/cww022. Epub 2016 Feb 24. Glycobiology. 2016. PMID: 26916883 Free PMC article. Review. - Thioglycosides Act as Metabolic Inhibitors of Bacterial Glycan Biosynthesis.
Quintana IL, Paul A, Chowdhury A, Moulton KD, Kulkarni SS, Dube DH. Quintana IL, et al. ACS Infect Dis. 2023 Oct 13;9(10):2025-2035. doi: 10.1021/acsinfecdis.3c00324. Epub 2023 Sep 12. ACS Infect Dis. 2023. PMID: 37698279 Free PMC article.
Cited by
- Chemoenzymatic Preparation of a Campylobacter jejuni Lipid-Linked Heptasaccharide on an Azide-Linked Polyisoprenoid.
Reid AJ, Erickson KM, Hazel JM, Lukose V, Troutman JM. Reid AJ, et al. ACS Omega. 2023 Apr 22;8(17):15790-15798. doi: 10.1021/acsomega.3c01657. eCollection 2023 May 2. ACS Omega. 2023. PMID: 37151508 Free PMC article. - HP0953 - hypothetical virulence factor overexpresion and localization during Helicobacter pylori infection of gastric epithelium.
Arteaga-Resendiz NK, Rodea GE, Ribas-Aparicio RM, Olivares-Cervantes AL, Castelán-Vega JA, Olivares-Trejo JJ, Mendoza-Elizalde S, López-Villegas EO, Colín C, Aguilar-Rodea P, Reyes-López A, Salazar García M, Velázquez-Guadarrama N. Arteaga-Resendiz NK, et al. World J Gastroenterol. 2022 Aug 7;28(29):3886-3902. doi: 10.3748/wjg.v28.i29.3886. World J Gastroenterol. 2022. PMID: 36157534 Free PMC article. - Development of a GalNAc-Tyrosine-Specific Monoclonal Antibody and Detection of Tyrosine _O_-GalNAcylation in Numerous Human Tissues and Cell Lines.
Xia L, Bellomo TR, Gibadullin R, Congdon MD, Edmondson EF, Li M, Wlodawer A, Li C, Temme JS, Patel P, Butcher D, Gildersleeve JC. Xia L, et al. J Am Chem Soc. 2022 Sep 14;144(36):16410-16422. doi: 10.1021/jacs.2c04477. Epub 2022 Sep 2. J Am Chem Soc. 2022. PMID: 36054098 Free PMC article. - Synthesis and Application of Rare Deoxy Amino l-Sugar Analogues to Probe Glycans in Pathogenic Bacteria.
Luong P, Ghosh A, Moulton KD, Kulkarni SS, Dube DH. Luong P, et al. ACS Infect Dis. 2022 Apr 8;8(4):889-900. doi: 10.1021/acsinfecdis.2c00060. Epub 2022 Mar 18. ACS Infect Dis. 2022. PMID: 35302355 Free PMC article. - Selective Exoenzymatic Labeling of Lipooligosaccharides of Neisseria gonorrhoeae with α2,6-Sialoside Analogues.
de Jong H, Moure MJ, Hartman JEM, Bosman GP, Ong JY, Bardoel BW, Boons GJ, Wösten MMSM, Wennekes T. de Jong H, et al. Chembiochem. 2022 Oct 6;23(19):e202200340. doi: 10.1002/cbic.202200340. Epub 2022 Aug 23. Chembiochem. 2022. PMID: 35877976 Free PMC article.
References
- Dube DH; Champasa K; Wang B, Chemical tools to discover and target bacterial glycoproteins. Chem Commun 2011, 47 (1), 87–101. - PubMed
- Edens RE, Polysaccharides: structural diversity and functional versatility; Dumitriu S, Ed.; Marcel Dekker: New York. 2005.
- Cigana C; Curcurù L; Leone MR; Ieranò T; Lorè NI; Bianconi I; Silipo A; Cozzolino F; Lanzetta R; Molinaro A; Bernardini ML; Bragonzi A, Pseudomonas aeruginosa exploits lipid A and muropeptides modification as a strategy to lower innate immunity during cystic fibrosis lung infection. PLoS One 2009, 4 (12), e8439. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources