AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence - PubMed (original) (raw)
. 2021 Jun 16;11(1):12728.
doi: 10.1038/s41598-021-91456-0.
Vyacheslav Brover 2, Narjol Gonzalez-Escalona 3, Jonathan G Frye 4, Julie Haendiges 3, Daniel H Haft 2, Maria Hoffmann 3, James B Pettengill 3, Arjun B Prasad 2, Glenn E Tillman 5, Gregory H Tyson 6, William Klimke 2
Affiliations
- PMID: 34135355
- PMCID: PMC8208984
- DOI: 10.1038/s41598-021-91456-0
AMRFinderPlus and the Reference Gene Catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence
Michael Feldgarden et al. Sci Rep. 2021.
Abstract
Antimicrobial resistance (AMR) is a significant public health threat. With the rise of affordable whole genome sequencing, in silico approaches to assessing AMR gene content can be used to detect known resistance mechanisms and potentially identify novel mechanisms. To enable accurate assessment of AMR gene content, as part of a multi-agency collaboration, NCBI developed a comprehensive AMR gene database, the Bacterial Antimicrobial Resistance Reference Gene Database and the AMR gene detection tool AMRFinder. Here, we describe the expansion of the Reference Gene Database, now called the Reference Gene Catalog, to include putative acid, biocide, metal, stress resistance genes, in addition to virulence genes and species-specific point mutations. Genes and point mutations are classified by broad functions, as well as more detailed functions. As we have expanded both the functional repertoire of identified genes and functionality, NCBI released a new version of AMRFinder, known as AMRFinderPlus. This new tool allows users the option to utilize only the core set of AMR elements, or include stress response and virulence genes, too. AMRFinderPlus can detect acquired genes and point mutations in both protein and nucleotide sequence. In addition, the evidence used to identify the gene has been expanded to include whether nucleotide or protein sequence was used, its location in the contig, and presence of an internal stop codon. These database improvements and functional expansions will enable increased precision in identifying AMR genes, linking AMR genotypes and phenotypes, and determining possible relationships between AMR, virulence, and stress response.
Conflict of interest statement
The authors declare no competing interests.
Figures
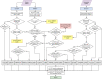
Figure 1
Overview of AMRFinderPlus gene identification method using protein and nucleotide sequences. When AMRFinderPlus is run in protein-only or nucleotide-only modes only the relevant side of the flowchart above is utilized. Note that HMMs are not used with nucleotide-only data which results in reduced sensitivity for some families. "Ignored" matches are not used, however an alternative method may still report the protein (e.g., an ignored HMM match will not suppress reporting based on BLASTP or BLASTX hit). *Details are simplified in this diagram; a more detailed description is included in the methods text.
Similar articles
- Distribution of Antimicrobial Resistance and Virulence Genes within the Prophage-Associated Regions in Nosocomial Pathogens.
Kondo K, Kawano M, Sugai M. Kondo K, et al. mSphere. 2021 Aug 25;6(4):e0045221. doi: 10.1128/mSphere.00452-21. Epub 2021 Jul 7. mSphere. 2021. PMID: 34232073 Free PMC article. - Keeping up with the pathogens: improved antimicrobial resistance detection and prediction from Pseudomonas aeruginosa genomes.
Madden DE, Baird T, Bell SC, McCarthy KL, Price EP, Sarovich DS. Madden DE, et al. Genome Med. 2024 Jun 7;16(1):78. doi: 10.1186/s13073-024-01346-z. Genome Med. 2024. PMID: 38849863 Free PMC article. - Genome analysis of antimicrobial resistance, virulence, and plasmid presence in Turkish Salmonella serovar Infantis isolates.
Acar S, Bulut E, Stasiewicz MJ, Soyer Y. Acar S, et al. Int J Food Microbiol. 2019 Oct 16;307:108275. doi: 10.1016/j.ijfoodmicro.2019.108275. Epub 2019 Jul 27. Int J Food Microbiol. 2019. PMID: 31408739 - Twenty-first century molecular methods for analyzing antimicrobial resistance in surface waters to support One Health assessments.
Franklin AM, Brinkman NE, Jahne MA, Keely SP. Franklin AM, et al. J Microbiol Methods. 2021 May;184:106174. doi: 10.1016/j.mimet.2021.106174. Epub 2021 Mar 24. J Microbiol Methods. 2021. PMID: 33774111 Free PMC article. Review. - Molecular Methods for Detection of Antimicrobial Resistance.
Anjum MF, Zankari E, Hasman H. Anjum MF, et al. Microbiol Spectr. 2017 Dec;5(6):10.1128/microbiolspec.arba-0011-2017. doi: 10.1128/microbiolspec.ARBA-0011-2017. Microbiol Spectr. 2017. PMID: 29219107 Free PMC article. Review.
Cited by
- Genomic Characterization of 16S rRNA Methyltransferase-Producing Enterobacterales Reveals the Emergence of Klebsiella pneumoniae ST6260 Harboring rmtF, rmtB, _bla_NDM-5, _bla_OXA-232 and _bla_SFO-1 Genes in a Cancer Hospital in Bulgaria.
Sabtcheva S, Stoikov I, Georgieva S, Donchev D, Hodzhev Y, Dobreva E, Christova I, Ivanov IN. Sabtcheva S, et al. Antibiotics (Basel). 2024 Oct 10;13(10):950. doi: 10.3390/antibiotics13100950. Antibiotics (Basel). 2024. PMID: 39452216 Free PMC article. - Creating and leveraging bespoke large-scale knowledge graphs for comparative genomics and multi-omics drug discovery with SocialGene.
Clark CM, Kwan JC. Clark CM, et al. bioRxiv [Preprint]. 2024 Aug 19:2024.08.16.608329. doi: 10.1101/2024.08.16.608329. bioRxiv. 2024. PMID: 39229008 Free PMC article. Preprint. - Genomic Insights into the Microbial Agent Streptomyces albidoflavus MGMM6 for Various Biotechnology Applications.
Diabankana RGC, Frolov M, Keremli S, Validov SZ, Afordoanyi DM. Diabankana RGC, et al. Microorganisms. 2023 Nov 27;11(12):2872. doi: 10.3390/microorganisms11122872. Microorganisms. 2023. PMID: 38138016 Free PMC article. - Diversity of Clostridioides difficile PCR ribotypes isolated from freshwater sediments depends on the isolation method.
Rupnik M, Fuks A, Janezic S. Rupnik M, et al. Appl Environ Microbiol. 2024 Oct 23;90(10):e0144224. doi: 10.1128/aem.01442-24. Epub 2024 Sep 13. Appl Environ Microbiol. 2024. PMID: 39269162 - Phylogenomic investigation of an outbreak of fluoroquinolone-resistant Salmonella enterica subsp. enterica serovar Paratyphi A in Phnom Penh, Cambodia.
Dusadeepong R, Delvallez G, Cheng S, Meng S, Sreng N, Letchford J, Choun K, Teav S, Hardy L, Jacobs J, Hoang T, Seemann T, Howden BP, Glaser P, Stinear TP, Vandelannoote K. Dusadeepong R, et al. Microb Genom. 2023 Mar;9(3):mgen000972. doi: 10.1099/mgen.0.000972. Microb Genom. 2023. PMID: 36961484 Free PMC article.
References
- Do Nascimento, V. et al. Comparison of phenotypic and WGS-derived antimicrobial resistance profiles of enteroaggregative Escherichia coli isolated from cases of diarrhoeal disease in England, 2015–16. J. Antimicrob. Chemother.72, 3288–3297. 10.1093/jac/dkx301 (2017). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials