Phenolic compounds disrupt spike-mediated receptor-binding and entry of SARS-CoV-2 pseudo-virions - PubMed (original) (raw)
Phenolic compounds disrupt spike-mediated receptor-binding and entry of SARS-CoV-2 pseudo-virions
Anna Goc et al. PLoS One. 2021.
Abstract
In the pursuit of suitable and effective solutions to SARS-CoV-2 infection, we investigated the efficacy of several phenolic compounds in controlling key cellular mechanisms involved in its infectivity. The way the SARS-CoV-2 virus infects the cell is a complex process and comprises four main stages: attachment to the cognate receptor, cellular entry, replication and cellular egress. Since, this is a multi-part process, it creates many opportunities to develop effective interventions. Targeting binding of the virus to the host receptor in order to prevent its entry has been of particular interest. Here, we provide experimental evidence that, among 56 tested polyphenols, including plant extracts, brazilin, theaflavin-3,3'-digallate, and curcumin displayed the highest binding with the receptor-binding domain of spike protein, inhibiting viral attachment to the human angiotensin-converting enzyme 2 receptor, and thus cellular entry of pseudo-typed SARS-CoV-2 virions. Both, theaflavin-3,3'-digallate at 25 μg/ml and curcumin above 10 μg/ml concentration, showed binding with the angiotensin-converting enzyme 2 receptor reducing at the same time its activity in both cell-free and cell-based assays. Our study also demonstrates that brazilin and theaflavin-3,3'-digallate, and to a still greater extent, curcumin, decrease the activity of transmembrane serine protease 2 both in cell-free and cell-based assays. Similar pattern was observed with cathepsin L, although only theaflavin-3,3'-digallate showed a modest diminution of cathepsin L expression at protein level. Finally, each of these three compounds moderately increased endosomal/lysosomal pH. In conclusion, this study demonstrates pleiotropic anti-SARS-CoV-2 efficacy of specific polyphenols and their prospects for further scientific and clinical investigations.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Fig 1. Binding of RBD-spike protein of SARS-CoV-2 to human ACE2 receptor.
(A) Dose-dependent binding of RBD-SARS-CoV-2 to immobilized hACE2 receptor. Control– 0.025% DMSO, positive and negative controls were provided by the manufacturer; data are presented as % of control ± SD. (B) Dose-dependent binding of A546 cells expressing SARS-CoV-2 eGFP-spike protein, in the presence of indicated polyphenols at different concentrations, to soluble hACE2 receptor. Control– 0.25% DMSO; positive and negative controls were provided by the manufacturer; data are presented as % of control ± SD. (C) Viability of A549 cells, positive control—100% dead cells, negative control—addition-free sample; TF-3—theaflavin-3,3’-digallate; # p ≤ 0.05, Δ p ≤ 0.01, * p ≤ 0.001.
Fig 2. Binding of SARS-CoV-2 pseudo-virion to human ACE2 receptor.
(A) Dose-dependent binding of SARS-CoV-2 spike protein-encapsulated pseudo-virions to A549 cells stably overexpressing human ACE2 receptor evaluated after 1h incubation. (B) Dose-dependent binding of SARS-CoV-2 spike protein-encapsulated pseudo-virions to A549 cells stably overexpressing hACE2 receptor evaluated after 3h incubation. Data are presented as % of control ± SD; control– 0.025% DMSO, positive and negative controls were provided by the manufacturer; TF-3 –theaflavin-3,3’-digallate # p ≤ 0.05, Δ p ≤ 0.01, * p ≤ 0.001.
Fig 3. SARS-CoV-2 eGFP-luciferase-pseudo-virion cellular entry.
(A) Attachment and entry of SARS-CoV-2 pseudo-virions with encapsulated eGFP-luciferase spike protein was evaluated without spinfection after 48h incubation. (B) Attachment and entry of SARS-CoV-2 pseudo-virions with encapsulated eGFP-luciferase spike protein was evaluated with spinfection after 48h incubation. Data are presented as % of control ± SD; TF-3 –theaflavin-3,3’-digallate # p ≤ 0.05, Δ p ≤ 0.01, * p ≤ 0.001. Control– 0.025% DMSO, positive control—bald SARS-CoV-2 eGFP-luciferase-pseudo-virions, negative control—ΔG-luciferase rVSV pseudo-typed particles; red fame—concentrations that showed 85–100% cytotoxicity.
Fig 4. Effect of selected polyphenols on fusion to human ACE2 receptor overexpressing A549 cells.
(A) Cell-cell fusion of A549 cells expressing eGFP spike protein with A549 cells stably expressing human ACE2 receptor. The scale bar indicates 250 μm. (B) Quantitative analysis of formed syncytia. Experiments were done in triplicate and repeated three times. Data are presented as % of control ± SD; TF-3 –theaflavin-3,3’-digallate Δ p ≤ 0.01, * p ≤ 0.001. Control– 0.025% DMSO, positive control– 20 μg/ml anti-ACE2 antibody.
Fig 5. Effects of selected polyphenols on cellular membrane associated proteases.
(A) Binding of indicated polyphenols at different concentrations to hACE2 receptor. Data are presented as % of control ± SD; control– 0.025% DMSO, positive control– 50% DMSO. (B) Activity of recombinant hACE2 upon treatment with indicated polyphenols at different concentrations. (left panel). Activity of cellular hACE2 upon treatment with indicated polyphenols at different concentrations. (right panel). Data are presented as % of control ± SD; * p ≤ 0.001. Control– 0.025% DMSO, positive control– 10% DMSO. (C) Activity of recombinant TMPTSS2 upon treatment with indicated polyphenols at different. (left panel). Activity of cellular TMPTSS2 upon treatment with indicated polyphenols at different concentrations (right panel). Data are presented as % of control ± SD; # p ≤ 0.05, Δ p ≤ 0.01, * p ≤ 0.001. Control– 0.025% DMSO, positive control– 50–100 μM camostat mesylate. (D) Western blot analysis of hACE2 and TMPRSS2 expression in A549 cells upon treatment with indicated polyphenols with different concentration for 48h period. Data are presented as % of control ± SD; control– 0.025% DMSO, TF-3 –theaflavin-3,3’-digallate.
Fig 6. Effect of selected polyphenols on cathepsin L.
(A) Activity of purified cathepsin L enzyme upon treatment with indicated polyphenols at different concentrations (left panel). Activity of cellular cathepsin L upon treatment with indicated polyphenols at different concentrations (right panel). Data are presented as % of control ± SD; Δ p ≤ 0.01, * p ≤ 0.001, + p < 0.054. Control– 0.025% DMSO, positive control– 0.1 μM E-64. (B) Western blot analysis of cathepsin L expression in A549 cells treated with indicated polyphenols with different concentration for 24h. (left panel) and quantified as band densitometry analysis indicating changes in protein expression (right panel). Data are presented as % of control ± SD; control– 0.025% DMSO, TF-3 –theaflavin-3,3’-digallate.
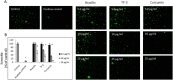
Fig 7. Effect of selected polyphenols on internal pH and endosome acidification.
(A) Intracellular/lysosomal pH measurement. pHrodo™ Green AM dye and additional incubation for 30 min. at 37°C. Cells were then washed and fluorescence was measured at Ex/Em = 535/595 nm. Intracellular pH identification was done using standard curve prepared by measuring fluorescence in the presence of standard buffers with indicated pH as described in Material and Methods section. (B) Endosomal pH measurement in A549 cells treated with indicated polyphenols at different concentrations for 3h at 37°C. Scale bar indicates 50 μm. Images are representative of all observed fields. Experiments were done in triplicates and repeated three times. Data are presented as % of control ± SD. TF-3 –theaflavin-3,3’-digallate; control– 0.025% DMSO, positive control– 20 mM ammonia chloride.
Similar articles
- The Inhibiting Effect of GB-2, (+)-Catechin, Theaflavin, and Theaflavin 3-Gallate on Interaction between ACE2 and SARS-CoV-2 EG.5.1 and HV.1 Variants.
Lu CK, Lung J, Shu LH, Liu HT, Wu YH, Lin YS, Yang YH, Wu YH, Wu CY. Lu CK, et al. Int J Mol Sci. 2024 Aug 31;25(17):9498. doi: 10.3390/ijms25179498. Int J Mol Sci. 2024. PMID: 39273444 Free PMC article. - Targeting the viral-entry facilitators of SARS-CoV-2 as a therapeutic strategy in COVID-19.
Muralidar S, Gopal G, Visaga Ambi S. Muralidar S, et al. J Med Virol. 2021 Sep;93(9):5260-5276. doi: 10.1002/jmv.27019. Epub 2021 May 3. J Med Virol. 2021. PMID: 33851732 Free PMC article. Review. - SARS-CoV-2 infects cells after viral entry via clathrin-mediated endocytosis.
Bayati A, Kumar R, Francis V, McPherson PS. Bayati A, et al. J Biol Chem. 2021 Jan-Jun;296:100306. doi: 10.1016/j.jbc.2021.100306. Epub 2021 Jan 19. J Biol Chem. 2021. PMID: 33476648 Free PMC article. - Polyunsaturated ω-3 fatty acids inhibit ACE2-controlled SARS-CoV-2 binding and cellular entry.
Goc A, Niedzwiecki A, Rath M. Goc A, et al. Sci Rep. 2021 Mar 4;11(1):5207. doi: 10.1038/s41598-021-84850-1. Sci Rep. 2021. PMID: 33664446 Free PMC article. - Defining diverse spike-receptor interactions involved in SARS-CoV-2 entry: Mechanisms and therapeutic opportunities.
Anderson M, Lopez J, Wyr M, Ramirez PW. Anderson M, et al. Virology. 2025 Jun;607:110507. doi: 10.1016/j.virol.2025.110507. Epub 2025 Mar 21. Virology. 2025. PMID: 40157321 Review.
Cited by
- Curcumin as an antiviral agent and immune-inflammatory modulator in COVID-19: A scientometric analysis.
Liu K, Zhu Y, Cao X, Liu Y, Ying R, Huang Q, Gao P, Zhang C. Liu K, et al. Heliyon. 2023 Nov 2;9(11):e21648. doi: 10.1016/j.heliyon.2023.e21648. eCollection 2023 Nov. Heliyon. 2023. PMID: 38027776 Free PMC article. - 2,4-Dihydroxycinnamic acid as spike ACE2 inhibitor and apigenin as RdRp inhibitor in Nimbamritadi Panchatiktam Kashayam against COVID-19: an in silico and in vitro approach.
Murali M, Nair B, Vishnu VR, Aneesh TP, Nath LR. Murali M, et al. Mol Divers. 2023 Oct;27(5):2353-2363. doi: 10.1007/s11030-022-10552-z. Epub 2022 Nov 10. Mol Divers. 2023. PMID: 36357813 Free PMC article. - Polyphenols: From Theory to Practice.
Bertelli A, Biagi M, Corsini M, Baini G, Cappellucci G, Miraldi E. Bertelli A, et al. Foods. 2021 Oct 27;10(11):2595. doi: 10.3390/foods10112595. Foods. 2021. PMID: 34828876 Free PMC article. Review. - Optimization of Anti-SARS-CoV-2 Treatments Based on Curcumin, Used Alone or Employed as a Photosensitizer.
Zupin L, Fontana F, Clemente L, Borelli V, Ricci G, Ruscio M, Crovella S. Zupin L, et al. Viruses. 2022 Sep 27;14(10):2132. doi: 10.3390/v14102132. Viruses. 2022. PMID: 36298687 Free PMC article. - Molecular Mechanisms of Possible Action of Phenolic Compounds in COVID-19 Protection and Prevention.
Gligorijevic N, Radomirovic M, Nedic O, Stojadinovic M, Khulal U, Stanic-Vucinic D, Cirkovic Velickovic T. Gligorijevic N, et al. Int J Mol Sci. 2021 Nov 17;22(22):12385. doi: 10.3390/ijms222212385. Int J Mol Sci. 2021. PMID: 34830267 Free PMC article. Review.
References
- World Health Organization. Coronavirus disease (COVID-19). 2020. [cited 2021 January 28]. https://covid19.who.int.
- International Committee on Taxonomy of Viruses. Severe acute respiratory syndrome-related coronavirus. 2020. [cited 2021 January 28]. https://talk.ictvonline.org/taxonomy.
- Masters PS, Perlman S. Fields Virology. In: Knipe DM, Howley PM, editors. Philadelphia, PA: Lippincott William and Wilkins; 2013. Pp. 825–858.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous