The Progenetix oncogenomic resource in 2021 - PubMed (original) (raw)
Meta-Analysis
The Progenetix oncogenomic resource in 2021
Qingyao Huang et al. Database (Oxford). 2021.
Abstract
In cancer, copy number aberrations (CNAs) represent a type of nearly ubiquitous and frequently extensive structural genome variations. To disentangle the molecular mechanisms underlying tumorigenesis as well as identify and characterize molecular subtypes, the comparative and meta-analysis of large genomic variant collections can be of immense importance. Over the last decades, cancer genomic profiling projects have resulted in a large amount of somatic genome variation profiles, however segregated in a multitude of individual studies and datasets. The Progenetix project, initiated in 2001, curates individual cancer CNA profiles and associated metadata from published oncogenomic studies and data repositories with the aim to empower integrative analyses spanning all different cancer biologies. During the last few years, the fields of genomics and cancer research have seen significant advancement in terms of molecular genetics technology, disease concepts, data standard harmonization as well as data availability, in an increasingly structured and systematic manner. For the Progenetix resource, continuous data integration, curation and maintenance have resulted in the most comprehensive representation of cancer genome CNA profiling data with 138 663 (including 115 357 tumor) copy number variation (CNV) profiles. In this article, we report a 4.5-fold increase in sample number since 2013, improvements in data quality, ontology representation with a CNV landscape summary over 51 distinctive National Cancer Institute Thesaurus cancer terms as well as updates in database schemas, and data access including new web front-end and programmatic data access. Database URL: progenetix.org.
© The Author(s) 2021. Published by Oxford University Press.
Figures
Figure 1.
The currently available CNA data points in Progenetix and TCGA Progenetix database contain 115 357 cancer samples with 92 307 mapped to the 51 defined critical nodes in NCIt ontology tree and 23 050 samples not mapped to the tree (black), whereas TCGA repository contains 11 090 samples with 9103 samples mapped and 1987 samples not mapped to the tree (black). Colors of the stacked bar plot (left) match the branch colors on NCIt ontology tree (right).
Figure 4.
Demonstration of further functionality pages: A. Publication search; B. NCIt hierarchical tree navigation A: Cancer-genomics-associated publications are recorded with number of samples stratified by technology used. The publications can be filtered by keywords; B: Part of the sample subsets contained in Progenetix under the hierarchical NCIt classification tree. It allows for selection of sample subsets at different levels; C: User can upload custom segment files for data visualization.
Figure 2.
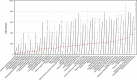
The genomic CNV fraction across 51 NCIt umbrella nodes Each dot represents one sample’s CNV fraction range from 0 to 1 and the red horizontal line indicates median CNV of the respective cancer type. Each cancer type contains between 104 and 11 804 CNV profiles (median 904; See Supplementary Table S1).
Figure 3.
Beacon-style query using fuzzy ranges to identify biosamples with variants matching the CNA range This example queries for a continuous, focal duplication covering the complete MYC gene’s coding region with < = 6 Mb in size. A: Filter for dataset; B: filter for cancer classification (NCIt and ICD-O-3 ontology terms available); C: additional filter, e.g. Cellosaurus; D: additional filter for geographic location; E: external link to UCSC browser to view the alignment of matched variants; F: cancer type classification sorted by frequency of the matched biosamples present in the subset; G: list of matched biosamples with description, statistics and reference. More detailed biosample information can be viewed through ‘id’ link to the sample detail page; H: matched variants with reference to biosamples can be downloaded in json or csv format.
Similar articles
- Progenetix: 12 years of oncogenomic data curation.
Cai H, Kumar N, Ai N, Gupta S, Rath P, Baudis M. Cai H, et al. Nucleic Acids Res. 2014 Jan;42(Database issue):D1055-62. doi: 10.1093/nar/gkt1108. Epub 2013 Nov 12. Nucleic Acids Res. 2014. PMID: 24225322 Free PMC article. - CNApp, a tool for the quantification of copy number alterations and integrative analysis revealing clinical implications.
Franch-Expósito S, Bassaganyas L, Vila-Casadesús M, Hernández-Illán E, Esteban-Fabró R, Díaz-Gay M, Lozano JJ, Castells A, Llovet JM, Castellví-Bel S, Camps J. Franch-Expósito S, et al. Elife. 2020 Jan 15;9:e50267. doi: 10.7554/eLife.50267. Elife. 2020. PMID: 31939734 Free PMC article. - CNVIntegrate: the first multi-ethnic database for identifying copy number variations associated with cancer.
Chattopadhyay A, Teoh ZH, Wu CY, Juang JJ, Lai LC, Tsai MH, Wu CH, Lu TP, Chuang EY. Chattopadhyay A, et al. Database (Oxford). 2021 Jul 14;2021:baab044. doi: 10.1093/database/baab044. Database (Oxford). 2021. PMID: 34259866 Free PMC article. - Mountains and Chasms: Surveying the Oncogenomic Publication Landscape.
Carrio-Cordo P, Baudis M. Carrio-Cordo P, et al. Oncology. 2020;98(6):332-343. doi: 10.1159/000493192. Epub 2018 Oct 26. Oncology. 2020. PMID: 30368507 Review. - Oncogenomic portals for the visualization and analysis of genome-wide cancer data.
Klonowska K, Czubak K, Wojciechowska M, Handschuh L, Zmienko A, Figlerowicz M, Dams-Kozlowska H, Kozlowski P. Klonowska K, et al. Oncotarget. 2016 Jan 5;7(1):176-92. doi: 10.18632/oncotarget.6128. Oncotarget. 2016. PMID: 26484415 Free PMC article. Review.
Cited by
- Breast tumors with elevated expression of 1q candidate genes confer poor clinical outcome and sensitivity to Ras/PI3K inhibition.
Muthuswami M, Ramesh V, Banerjee S, Viveka Thangaraj S, Periasamy J, Bhaskar Rao D, Barnabas GD, Raghavan S, Ganesan K. Muthuswami M, et al. PLoS One. 2013 Oct 17;8(10):e77553. doi: 10.1371/journal.pone.0077553. eCollection 2013. PLoS One. 2013. PMID: 24147022 Free PMC article. - Proof of concept: prognostic value of the plasmatic concentration of circulating cell free DNA in desmoid tumors using ddPCR.
Macagno N, Fina F, Penel N, Bouvier C, Nanni I, Duffaud F, Rouah R, Lacarelle B, Ouafik L, Bonvalot S, Salas S. Macagno N, et al. Oncotarget. 2018 Apr 6;9(26):18296-18308. doi: 10.18632/oncotarget.24817. eCollection 2018 Apr 6. Oncotarget. 2018. PMID: 29719606 Free PMC article. - cancercelllines.org-a novel resource for genomic variants in cancer cell lines.
Paloots R, Baudis M. Paloots R, et al. Database (Oxford). 2024 Apr 30;2024:baae030. doi: 10.1093/database/baae030. Database (Oxford). 2024. PMID: 38687868 Free PMC article. - CYP2D6 copy number determination using digital PCR.
Wang WY, Lin L, Boone EC, Stevens J, Gaedigk A. Wang WY, et al. Front Pharmacol. 2024 Aug 14;15:1429286. doi: 10.3389/fphar.2024.1429286. eCollection 2024. Front Pharmacol. 2024. PMID: 39206265 Free PMC article. - labelSeg: segment annotation for tumor copy number alteration profiles.
Zhao H, Baudis M. Zhao H, et al. Brief Bioinform. 2024 Jan 22;25(2):bbad541. doi: 10.1093/bib/bbad541. Brief Bioinform. 2024. PMID: 38300514 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources