Dual-Specificity Phosphatase 9 Regulates Cellular Proliferation and Predicts Recurrence After Surgery in Hepatocellular Carcinoma - PubMed (original) (raw)
. 2021 Mar 11;5(7):1310-1328.
doi: 10.1002/hep4.1701. eCollection 2021 Jul.
Affiliations
- PMID: 34278178
- PMCID: PMC8279460
- DOI: 10.1002/hep4.1701
Dual-Specificity Phosphatase 9 Regulates Cellular Proliferation and Predicts Recurrence After Surgery in Hepatocellular Carcinoma
Kui Chen et al. Hepatol Commun. 2021.
Abstract
Hepatocellular carcinoma (CC) is a common and deadly cancer with complex molecular pathogenesis. Little is known about dual-specificity phosphatases (DUSPs) in HCC. We investigated DUSP9 expression in human HCC, associations between DUSP9 and patient outcomes, and effects of altered DUSP9 expression on HCC biology. We studied public data sets as well as 196 patients at our institution who had HCC resections. Quantitative real-time reverse transcription polymerase chain reaction and western blot demonstrated that DUSP9 expression was increased >10-fold in HCC compared to adjacent liver and healthy controls (P = 0.005). Kaplan-Meier and multivariable regression analyses revealed that higher DUSP9 expression was associated with shorter disease-free survival (high DUSP9, 1.6; 95% confidence interval, 0.9-2.3 vs. low DUSP9, 3.4; 95% confidence interval, 1.8-5.0 years; P = 0.04) and increased risk of recurrence (hazard ratio 1.55; 95% confidence interval, 1.01-2.67; P = 0.05) after resection. DUSP9 complementary DNA (cDNA) was cloned using rapid amplification of cDNA ends, revealing two DUSP9 isoforms in human HCC cells. Studies of transcriptional regulation using promoter-luciferase reporter constructs suggested that DUSP9 transcription is regulated by E26 transformation-specific transcription factors. Proliferation of hepatic cells in vitro was enhanced by lentiviral-mediated overexpression of DUSP9. In contrast, DUSP9 knockout HCC cells generated using clustered regularly interspaced short palindromic repeats (CRISPR) demonstrated decreased HCC proliferation and doxorubicin resistance in vitro and impaired xenograft growth in vivo. RNA sequencing, gene set enrichment, and network/pathway analysis revealed that DUSP9 knockout is associated with activation of protein kinase activity and apoptosis. Conclusion: DUSP9 regulates cell proliferation and predicts recurrence after surgery in HCC. DUSP9 may represent a novel prognostic candidate and therapeutic target. Additional studies are warranted to further explore the role and regulation of DUSP9 in HCC.
© 2021 The Authors. Hepatology Communications published by Wiley Periodicals LLC on behalf of the American Association for the Study of Liver Diseases.
Figures
FIG. 1
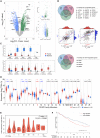
DUSP9 expression in HCC in public data sets. (A) Volcano plot of gene expression data from
GSE1133
(left), NCI60_U133A_CEL (right top), and
GSE25097
. (B) Venn diagrams showing up‐regulated (top) and down‐regulated (bottom) genes shared by all three comparisons in (A). Correlation plots (middle) of DUSP9, AFP, and GPC3 expression in TCGA data. (C) Box plot of DUSP9 expression in different cancers; horizontal bars indicate the median, box indicates interquartile range from Q1 to Q3 with jitter size of 0.4; *P < 0.05. (D) Violin plot showing expression of DUSPs in HCC; violin plots indicate probability density and horizontal bars indicate the median; significant mean fold changes of gene expression in tumor versus adjacent are indicated below asterisks; ****P < 0.0001, ***P < 0.001, **P < 0.01, *P < 0.05. (E) Violin plot of DUSP9 expression in progressive stages of HCC; white dots indicate median, vertical black boxes indicate interquartile range from Q1 to Q3. (F) Kaplan‐Meier analysis of DFS following HCC resection in TCGA cohort according to DUSP9 expression. Abbreviations: BLCA, bladder cancer; CD81, cluster of differentiation 81; CESC, cervical cancer; COL2A1, collagen type 2 α1; DLK1, delta like non‐canonical Notch ligand 1; ERBB3, erb‐b2 receptor tyrosine kinase 3; FC, fold change; FLiver, fetal liver; GBM, glioblastoma; HNSC, head‐neck squamous cell carcinoma; KICH/KIRC/KIRP, kidney cancers: chromophobe renal cell carcinoma/clear cell renal cell carcinoma/papillary renal cell carcinoma; LAML, acute myeloid leukemia; LIHC, liver hepatocellular carcinoma; LUSC, lung squamous cell carcinoma; RPS6KA3, ribosomal protein S6 kinase A3; THYM, thymoma; TIMP1, tissue inhibitor of metalloproteinase 1; TPM, transcripts per million; UCS, uterine cancer.
FIG. 2
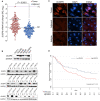
DUSP9 expression in HCC in patients in the UHN. (A) Dot plot of qPCR (using primer pair fp2/rp2) for DUSP9 expression in HCC tumors (red) and adjacent liver tissues (blue). Horizontal black bar indicates mean, vertical black bar indicates SEM. (B) Western blot of DUSP9 in hepatic cell lines, HCC tumors, and adjacent liver. (C) Immunofluorescence staining of human HCC tumor and adjacent liver. White scale bars, 15 μm; yellow scale bar, 10 μm. (D) Kaplan‐Meier analysis of DFS following HCC resection in UHN cohort according to DUSP9 expression. Abbreviations: DAPI, 4´,6‐diamidino‐2‐phenylindole; FL, fetal liver; N, adjacent liver; NC, normal control; RQ, relative quantification; T, hepatocellular carcinoma tumors.
FIG. 3
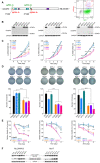
Identification of HCC‐specific DUSP9 isoform. (A) Schematic illustration comparing WT DUSP9 (NM_001395.4 at top) and N‐terminal truncated DUSP9 isoform (bottom, combination of MN308287 and BC034936.1). Black/green boxes indicate exons. Coding sequences are shown with key functional domains. Green segments in truncated isoform represent sequence differences compared to WT DUSP9. Primer pairs (fp1/rp1, fp2/rp2) are also indicated. (B) Combination of 5’RACE‐PCR amplified fragment (MN) with published partial sequence BC034936.1. MN308287 consists of g1 to t674, including 57 nucleotides of overlap with BC034936.1 (white letters), revealing 617 nucleotides of novel 5’ sequence. Red “atg” represents the predicted translation initiation site of the HCC‐specific DUSP9 isoform. (C) qPCR using primers selective for the truncated DUSP9 isoform (fp1/rp1, blue bars) or all DUSP9 isoforms (fp2/rp2, red bars) in HCC cell lines (RQ normalized to DUSP9 mRNA level in THLE‐2). Graph shows mean + SEM. (D) qPCR using primers selective for the truncated DUSP9 isoform (fp1/rp1, blue box) or all DUSP9 isoforms (fp2/rp2, red box) in 10 HCC tumors and adjacent liver tissues. Graph shows IQR (box), median (horizontal line), and outliers (whiskers). (E) Western blot of DUSP9 in four HCC cell lines, human HCC tumors, and adjacent liver. Abbreviations: aa, amino acid; Cdc25, cell division cycle 25; chr, chromosome; FC, fold change; N, adjacent liver; RQ, relative quantification; T, HCC tumors.
FIG. 4
Characterization of DUSP9 KO HCC cells. (A) Targeting strategy for CRISPR‐mediated DUSP9 KO. Cas9n‐mediated DNA cleavage sites were designed after the first start codon (Hep3B and PP5 cells, using sgRNA‐1a/1b) or after the second start codon (HepG2 and Huh7 cells, using sgRNA‐2a/2b). FACS plot (right) shows gating of transfected cells with dual fluorescent tags (GFP for sgRNA‐1a or 2a, mCherry for sgRNA‐1b or 2b) for further screening. (B) Western blots for DUSP9 in WT and KO cell lines. (C) WST‐1 assay on DUSP9 WT and KO cells. Data represent mean of six replicates per group; ****P < 0.0001. (D) Colony formation assays on DUSP9 KO and WT cells. Data show mean + SEM of three replicates per group; ****P < 0.0001. (E) Relative viability assays of doxorubicin‐treated DUSP9 KO versus WT Hep3B (200 ng/mL), PP5 (400 ng/mL), and Huh7 (1 µg/mL) cells. Data show mean + SEM of six replicates per group; ****P < 0.0001. (F) Western blot of DUSP9 KO and WT cells for Erk1/2 (p44/42 MAPK) and phosphorylated Erk1/2 (phospho‐p44/42 MAPK). Abbreviation: FACS, fluorescence‐activated cell sorting.
FIG. 5
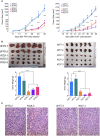
DUSP9 KO decreases human HCC xenograft growth in vivo. (A) Xenograft tumor growth curves generated by DUSP9 WT and KO HCC cells. (B) Xenografts generated by DUSP9 WT and KO HCC cells and corresponding bar graphs of tumor weights. Data in (A,B) show mean + SEM of eight replicates per group; ****P < 0.0001. (C) HCC xenografts generated by DUSP9 WT and KO HCC cells. Scale bar, 100 μm.
FIG. 6
RNA‐Seq analysis of DUSP9 WT and KO HCC cells. (A) Representative heatmap of expression clustering differentiates between DUSP9 KO and WT colonies across cell lines. DUSP9 KO is associated with decreased expression of 109 genes (top) and increased expression of 343 genes (bottom) (yellow, increased expression; blue, decreased expression). (B) Venn diagrams illustrating 343 up‐regulated and 109 down‐regulated genes consistently identified by pooled analysis of RNA‐Seq data from DUSP9 KO versus WT HCC cells using the R packages DESeq2, edgeR, and limma‐voom. (C) Clustering of 273 GO terms returned by Metascape functional enrichment analysis into five major groups. (D,E) Schematic illustrations of MAPK signaling and apoptosis pathways generated by pathview, highlighting specific genes with altered expression in association with DUSP9 KO (red, up‐regulated; green, down‐regulated). Abbreviations: AP1, activator protein 1; CACN, calcium voltage‐gated channel gene group; CASP8, caspase 8; Evi1, ecotropic viral integration site‐1; FLIP, FLICE‐inhibitory protein; GF, growth factor; HSP72, heat shock protein 72; PI3K, phosphoinositide 3‐kinase; TRAIL‐R, tumor necrosis factor–related apoptosis‐inducing ligand receptor.
Similar articles
- A novel prognostic biomarker SPC24 up-regulated in hepatocellular carcinoma.
Zhu P, Jin J, Liao Y, Li J, Yu XZ, Liao W, He S. Zhu P, et al. Oncotarget. 2015 Dec 1;6(38):41383-97. doi: 10.18632/oncotarget.5510. Oncotarget. 2015. PMID: 26515591 Free PMC article. - Overexpression of phosphatidylinositol 4-kinase type IIIα is associated with undifferentiated status and poor prognosis of human hepatocellular carcinoma.
Ilboudo A, Nault JC, Dubois-Pot-Schneider H, Corlu A, Zucman-Rossi J, Samson M, Le Seyec J. Ilboudo A, et al. BMC Cancer. 2014 Jan 6;14:7. doi: 10.1186/1471-2407-14-7. BMC Cancer. 2014. PMID: 24393405 Free PMC article. - Dual specificity phosphatase 9 (DUSP9) expression is down-regulated in the severe pre-eclamptic placenta.
Czikk MJ, Drewlo S, Baczyk D, Adamson SL, Kingdom J. Czikk MJ, et al. Placenta. 2013 Feb;34(2):174-81. doi: 10.1016/j.placenta.2012.11.029. Epub 2012 Dec 29. Placenta. 2013. PMID: 23276385 - Genome-scale CRISPR-Cas9 knockout screening in hepatocellular carcinoma with lenvatinib resistance.
Lu Y, Shen H, Huang W, He S, Chen J, Zhang D, Shen Y, Sun Y. Lu Y, et al. Cell Death Discov. 2021 Nov 18;7(1):359. doi: 10.1038/s41420-021-00747-y. Cell Death Discov. 2021. PMID: 34795217 Free PMC article. Review. - Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.
Frau M, Feo F, Pascale RM. Frau M, et al. J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
Cited by
- Identification and Validation of a Novel Pyroptosis-Related Gene Signature for Prognosis Prediction in Soft Tissue Sarcoma.
Qi L, Xu R, Wan L, Ren X, Zhang W, Zhang K, Tu C, Li Z. Qi L, et al. Front Genet. 2021 Dec 1;12:773373. doi: 10.3389/fgene.2021.773373. eCollection 2021. Front Genet. 2021. PMID: 34925457 Free PMC article. - Oncogenes and tumor suppressor genes: functions and roles in cancers.
Dakal TC, Dhabhai B, Pant A, Moar K, Chaudhary K, Yadav V, Ranga V, Sharma NK, Kumar A, Maurya PK, Maciaczyk J, Schmidt-Wolf IGH, Sharma A. Dakal TC, et al. MedComm (2020). 2024 May 31;5(6):e582. doi: 10.1002/mco2.582. eCollection 2024 Jun. MedComm (2020). 2024. PMID: 38827026 Free PMC article. Review. - Identifying specific TLS-associated genes as potential biomarkers for predicting prognosis and evaluating the efficacy of immunotherapy in soft tissue sarcoma.
Wang XX, Liu YP, Lu Y, Wu LH, Ren JY, Ji H, Wang X, Zhang HM. Wang XX, et al. Front Immunol. 2024 Apr 24;15:1372692. doi: 10.3389/fimmu.2024.1372692. eCollection 2024. Front Immunol. 2024. PMID: 38720884 Free PMC article. - DUSP9, a Dual-Specificity Phosphatase with a Key Role in Cell Biology and Human Diseases.
Khoubai FZ, Grosset CF. Khoubai FZ, et al. Int J Mol Sci. 2021 Oct 26;22(21):11538. doi: 10.3390/ijms222111538. Int J Mol Sci. 2021. PMID: 34768967 Free PMC article. Review. - miR-199a-5p targets DUSP14 to regulate cell proliferation, invasion and stemness in non-small cell lung cancer.
Zheng Y, Yang C, Xie S, Liu D, Wang H, Liu J. Zheng Y, et al. Heliyon. 2024 Apr 6;10(8):e29102. doi: 10.1016/j.heliyon.2024.e29102. eCollection 2024 Apr 30. Heliyon. 2024. PMID: 38644862 Free PMC article.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394‐424. - PubMed
- Villanueva A. Hepatocellular carcinoma. N Engl J Med 2019;380:1450‐1462. - PubMed
- Roayaie S, Jibara G, Tabrizian P, Park J‐W, Yang J, Yan L, et al. The role of hepatic resection in the treatment of hepatocellular cancer. Hepatology 2015;62:440‐451. - PubMed
- Sapisochin G, Goldaracena N, Laurence JM, Dib M, Barbas A, Ghanekar A, et al. The extended Toronto criteria for liver transplantation in patients with hepatocellular carcinoma: a prospective validation study. Hepatology 2016;64:2077‐2088. - PubMed
- Llovet JM, Ricci S, Mazzaferro V, Hilgard P, Gane E, Blanc J‐F, et al.; SHARP Investigators Study Group . Sorafenib in advanced hepatocellular carcinoma. N Engl J Med 2008;359:378‐390. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials