SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay - PubMed (original) (raw)
. 2021 Jul 30;7(31):eabh3409.
doi: 10.1126/sciadv.abh3409. Print 2021 Jul.
Saki Takahashi 2, Jill Hakim 2, J Daniel Kelly 3, Leonel Torres 2 4, Nikita S Iyer 4, Keirstinne Turcios 2, Owen Janson 2, Sadie E Munter 4, Cassandra Thanh 4, Joanna Donatelli 4, Christopher C Nixon 4, Rebecca Hoh 2, Viva Tai 2, Emily A Fehrman 2, Yanel Hernandez 2, Matthew A Spinelli 2, Monica Gandhi 2, Mary-Ann Palafox 5, Ana Vallari 5, Mary A Rodgers 5, John Prostko 5, John Hackett Jr 5, Lan Trinh 6, Terri Wrin 6, Christos J Petropoulos 6, Charles Y Chiu 7 8 9, Philip J Norris 10, Clara DiGermanio 10, Mars Stone 10, Michael P Busch 7 10, Susanna K Elledge 11, Xin X Zhou 11, James A Wells 11 12, Albert Shu 7, Theodore W Kurtz 7, John E Pak 13, Wesley Wu 13, Peter D Burbelo 14, Jeffrey I Cohen 15, Rachel L Rutishauser 4, Jeffrey N Martin 3, Steven G Deeks 2, Timothy J Henrich 4, Isabel Rodriguez-Barraquer 2, Bryan Greenhouse 2
Affiliations
- PMID: 34330709
- PMCID: PMC8324059
- DOI: 10.1126/sciadv.abh3409
SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay
Michael J Peluso et al. Sci Adv. 2021.
Abstract
Interpretation of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) serosurveillance studies is limited by poorly defined performance of antibody assays over time in individuals with different clinical presentations. We measured antibody responses in plasma samples from 128 individuals over 160 days using 14 assays. We found a consistent and strong effect of disease severity on antibody magnitude, driven by fever, cough, hospitalization, and oxygen requirement. Responses to spike protein versus nucleocapsid had consistently higher correlation with neutralization. Assays varied substantially in sensitivity during early convalescence and time to seroreversion. Variability was dramatic for individuals with mild infection, who had consistently lower antibody titers, with sensitivities at 6 months ranging from 33 to 98% for commercial assays. Thus, the ability to detect previous infection by SARS-CoV-2 is highly dependent on infection severity, timing, and the assay used. These findings have important implications for the design and interpretation of SARS-CoV-2 serosurveillance studies.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works. Distributed under a Creative Commons Attribution License 4.0 (CC BY).
Figures
Fig. 1. Longitudinal antibody kinetics.
Time since symptom onset is shown on the x axis versus the measured antibody response for each assay. For asymptomatic individuals, the time since the first positive polymerase chain reaction (PCR) test was used. Black points indicate individual time points, and longitudinal samples are connected with gray lines. y axes are transformed as indicated in Table 2. Assay units are as follows: S-Lum (conc, relative concentration), RBD-Lum (conc, relative concentration), RBD-LIPS (LU, light unit), RBD-Split Luc (RLU, relative light unit), S-Ortho IgG (S/C, sample result to calibrator result index), S-Ortho Ig (S/C, sample result to calibrator result index), S-DiaSorin (AU/ml, arbitrary unit per milliliter), N(full)-Lum (conc, relative concentration), N(frag)-Lum (conc, relative concentration), N-LIPS (LU, light unit), N-Split Luc (RLU, relative light unit), N-Abbott (S/C, sample result to calibrator result index), N-Roche (COI, cutoff index), and Neut-Monogram (ID50, 50% inhibitory dilution). Red dotted lines indicate cutoff values for positivity, as indicated in Table 2.
Fig. 2. Correlation of responses between assays.
(A) Spearman correlation of random intercepts derived from a mixed-effects model, representing responses at 21 days after symptom onset for each individual from the longitudinal data. Assays are sorted by hierarchical clustering using average distance clustering. Darker blue indicates higher correlation; colored label box indicates antigen for each binding assay and the neutralizing assay. (B) Pairwise scatterplots showing the random intercepts for the neutralizing assay (x axis) versus the random intercepts for each of the other assays (y axis). Assay units are indicated in Table 2.
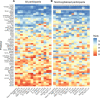
Fig. 3. Severity-stratified antibody response at baseline.
Swarm plot of antibody response at the baseline visit for each study participant by assay, stratified into individuals who experienced no symptoms, individuals who experienced symptoms but were not hospitalized, and those who experienced symptoms and were hospitalized. y axes are transformed as indicated in Table 2.
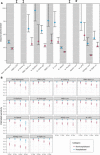
Fig. 4. Clinical predictors of antibody responses.
Rank of variable importance (1 = highest rank; 50 = lowest rank) in a random forest classifier of top half versus bottom half of responders for each assay, based on random intercepts, including (A) all individuals (n = 128) and (B) only individuals who were not hospitalized (n = 97), as hospitalization is a strong predictor of antibody response. Variable importance was determined as the reduction in classification error averaged across 10 runs of the algorithm. Variables only relevant to hospitalized individuals (i.e., whether the individual was hospitalized, whether oxygen was required, whether the individual was in the ICU, and whether the individual required a ventilator) were omitted from the classifier in (B) and shown in gray. In addition, HIV status is excluded as a predictor for the Neut-Monogram assay for reasons described in the main text. The dependent variable (individual-level random intercepts derived from a mixed-effects model) is dichotomized into “high” and “low,” determined by the random intercept being in the upper or lower half of all random intercepts for that assay, respectively. Full labels of the predictor variables are provided in table S10.
Fig. 5. Estimated time to seroreversion and assay sensitivity by time and hospitalization status.
(A) Mean time to seroreversion for individuals tested on each assay, stratified by hospitalization status, with 95% confidence intervals derived from bootstrapping. The symbol “‡” indicates increasing antibody responses over time (95% confidence interval for time to seroreversion was negative and did not cross 0), and the symbol “*” indicates antibody responses for which 95% confidence interval of time to seroreversion crossed 0. (B) Estimated sensitivity of each assay (showing posterior median estimates and 95% credible intervals), stratified by hospitalization status at 2-month intervals, from 0 to 6 months after seroconversion. Seroconversion was assumed to occur (if at all) 21 days after symptom onset (if symptomatic) or 21 days after positive PCR test (if asymptomatic).
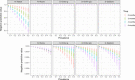
Fig. 6. Negative predictive values of the commercial assays.
Negative predictive values shown are based on the estimated assay sensitivities for nonhospitalized individuals in Fig. 5B, for a range of prevalence between 5 and 50% (x axis). Bottom panels show the same data with a smaller range in the y axis to visualize small differences.
Update of
- SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay.
Peluso MJ, Takahashi S, Hakim J, Kelly JD, Torres L, Iyer NS, Turcios K, Janson O, Munter SE, Thanh C, Nixon CC, Hoh R, Tai V, Fehrman EA, Hernandez Y, Spinelli MA, Gandhi M, Palafox MA, Vallari A, Rodgers MA, Prostko J, Hackett J Jr, Trinh L, Wrin T, Petroplolous CJ, Chiu CY, Norris PJ, DiGermanio C, Stone M, Busch MP, Elledge SK, Zhou XX, Wells JA, Shu A, Kurtz TW, Pak JE, Wu W, Burbelo PD, Cohen JI, Rutishauser RL, Martin JN, Deeks SG, Henrich TJ, Rodriguez-Barraquer I, Greenhouse B. Peluso MJ, et al. medRxiv [Preprint]. 2021 Mar 5:2021.03.03.21251639. doi: 10.1101/2021.03.03.21251639. medRxiv. 2021. PMID: 33688675 Free PMC article. Updated. Preprint.
Similar articles
- SARS-CoV-2 antibody magnitude and detectability are driven by disease severity, timing, and assay.
Peluso MJ, Takahashi S, Hakim J, Kelly JD, Torres L, Iyer NS, Turcios K, Janson O, Munter SE, Thanh C, Nixon CC, Hoh R, Tai V, Fehrman EA, Hernandez Y, Spinelli MA, Gandhi M, Palafox MA, Vallari A, Rodgers MA, Prostko J, Hackett J Jr, Trinh L, Wrin T, Petroplolous CJ, Chiu CY, Norris PJ, DiGermanio C, Stone M, Busch MP, Elledge SK, Zhou XX, Wells JA, Shu A, Kurtz TW, Pak JE, Wu W, Burbelo PD, Cohen JI, Rutishauser RL, Martin JN, Deeks SG, Henrich TJ, Rodriguez-Barraquer I, Greenhouse B. Peluso MJ, et al. medRxiv [Preprint]. 2021 Mar 5:2021.03.03.21251639. doi: 10.1101/2021.03.03.21251639. medRxiv. 2021. PMID: 33688675 Free PMC article. Updated. Preprint. - Wild-type SARS-CoV-2 neutralizing immunity decreases across variants and over time but correlates well with diagnostic testing.
O'Shea KM, Schuler CF 4th, Chen J, Troost JP, Wong PT, Chen K, O'Shea DR, Peng W, Gherasim C, Manthei DM, Valdez R, Baldwin JL, Baker JR Jr. O'Shea KM, et al. Front Immunol. 2023 Feb 8;14:1055429. doi: 10.3389/fimmu.2023.1055429. eCollection 2023. Front Immunol. 2023. PMID: 36845123 Free PMC article. - SARS-CoV-2 Antibody Testing in Health Care Workers: A Comparison of the Clinical Performance of Three Commercially Available Antibody Assays.
Allen N, Brady M, Carrion Martin AI, Domegan L, Walsh C, Houlihan E, Kerr C, Doherty L, King J, Doheny M, Griffin D, Molloy M, Dunne J, Crowley V, Holmes P, Keogh E, Naughton S, Kelly M, O'Rourke F, Lynagh Y, Crowley B, de Gascun C, Holder P, Bergin C, Fleming C, Ni Riain U, Conlon N; PRECISE Study Steering Group. Allen N, et al. Microbiol Spectr. 2021 Oct 31;9(2):e0039121. doi: 10.1128/Spectrum.00391-21. Epub 2021 Sep 29. Microbiol Spectr. 2021. PMID: 34585976 Free PMC article. - Performance of Elecsys Anti-SARS CoV-2 (Roche) and VIDAS Anti-SARS CoV-2 (Biomérieux) for SARS-CoV-2 Nucleocapsid and Spike Protein Antibody Detection.
Inés RM, Gabriela HTM, Paula CM, Magdalena TM, Jimena A, Salome KB, Javier AJ, Sebastián B, Lorena S, Adrián DL, Elisa R, Mauricio B, Tersita BM, Verónica GS, Beatriz IM. Inés RM, et al. EJIFCC. 2022 Aug 8;33(2):159-165. eCollection 2022 Aug. EJIFCC. 2022. PMID: 36313907 Free PMC article. Review. - Rapid, point-of-care antigen tests for diagnosis of SARS-CoV-2 infection.
Dinnes J, Sharma P, Berhane S, van Wyk SS, Nyaaba N, Domen J, Taylor M, Cunningham J, Davenport C, Dittrich S, Emperador D, Hooft L, Leeflang MM, McInnes MD, Spijker R, Verbakel JY, Takwoingi Y, Taylor-Phillips S, Van den Bruel A, Deeks JJ; Cochrane COVID-19 Diagnostic Test Accuracy Group. Dinnes J, et al. Cochrane Database Syst Rev. 2022 Jul 22;7(7):CD013705. doi: 10.1002/14651858.CD013705.pub3. Cochrane Database Syst Rev. 2022. PMID: 35866452 Free PMC article. Review.
Cited by
- Severe Acute Respiratory Syndrome Coronavirus 2 Seroprevalence and Reported Coronavirus Disease 2019 Cases in US Children, August 2020-May 2021.
Couture A, Lyons BC, Mehrotra ML, Sosa L, Ezike N, Ahmed FS, Brown CM, Yendell S, Azzam IA, Katić BJ, Cope A, Dickerson K, Stone J, Traxler LB, Dunn JR, Davis LB, Reed C, Clarke KEN, Flannery B, Charles MD. Couture A, et al. Open Forum Infect Dis. 2022 Jan 30;9(3):ofac044. doi: 10.1093/ofid/ofac044. eCollection 2022 Mar. Open Forum Infect Dis. 2022. PMID: 35198651 Free PMC article. - Association between COVID-19 and consistent mask wearing during contact with others outside the household-A nested case-control analysis, November 2020-October 2021.
Tjaden AH, Edelstein SL, Ahmed N, Calamari L, Dantuluri KL, Gibbs M, Hinkelman A, Mongraw-Chaffin M, Sanders JW, Saydah S, Plumb ID; COVID-19 Community Research Partnership Study Group. Tjaden AH, et al. Influenza Other Respir Viruses. 2023 Jan;17(1):e13080. doi: 10.1111/irv.13080. Epub 2023 Jan 5. Influenza Other Respir Viruses. 2023. PMID: 36606308 Free PMC article. - Seroprevalence of Antibodies to SARS-CoV-2 in Rural Households in Eastern Uganda, 2020-2022.
Briggs J, Takahashi S, Nayebare P, Cuu G, Rek J, Zedi M, Kizza T, Arinaitwe E, Nankabirwa JI, Kamya M, Jagannathan P, Jacobson K, Rosenthal PJ, Dorsey G, Greenhouse B, Ssewanyana I, Rodríguez-Barraquer I. Briggs J, et al. JAMA Netw Open. 2023 Feb 1;6(2):e2255978. doi: 10.1001/jamanetworkopen.2022.55978. JAMA Netw Open. 2023. PMID: 36790811 Free PMC article. - Prevalence of antibodies against SARS-CoV-2 in the Norwegian population, August 2021.
Tunheim G, Rø GØI, Chopra A, Aase A, Kran AB, Vaage JT, Lund-Johansen F, Hungnes O. Tunheim G, et al. Influenza Other Respir Viruses. 2022 Nov;16(6):1004-1013. doi: 10.1111/irv.13024. Epub 2022 Jun 30. Influenza Other Respir Viruses. 2022. PMID: 35770841 Free PMC article. - Pediatric SARS-CoV-2 Seroprevalence, Oregon, USA, November 1, 2020-June 30, 2022.
Falender RA, Mitchell PG, Guzman-Cottrill JA, Cieslak PR, Sutton M. Falender RA, et al. Emerg Infect Dis. 2023 Aug;29(8):1672-1675. doi: 10.3201/eid2908.230471. Emerg Infect Dis. 2023. PMID: 37486347 Free PMC article.
References
- Guan W.-J., Ni Z.-Y., Hu Y., Liang W.-H., Ou C.-Q., He J.-X., Liu L., Shan H., Lei C.-L., Hui D. S. C., Du B., Li L.-J., Zeng G., Yuen K.-Y., Chen R.-C., Tang C.-L., Wang T., Chen P.-Y., Xiang J., Li S.-Y., Wang J.-L., Liang Z.-J., Peng Y.-X., Wei L., Liu Y., Hu Y.-H., Peng P., Wang J.-M., Liu J.-Y., Chen Z., Li G., Zheng Z.-J., Qiu S.-Q., Luo J., Ye C.-J., Zhu S.-Y., Zhong N.-S.; China Medical Treatment Expert Group for Covid-19 , Clinical characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382, 1708–1720 (2020). - PMC - PubMed
- Richardson S., Hirsch J. S., Narasimhan M., Crawford J. M., McGinn T., Davidson K. W.; the Northwell COVID-19 Research Consortium, Barnaby D. P., Becker L. B., Chelico J. D., Cohen S. L., Cookingham J., Coppa K., Diefenbach M. A., Dominello A. J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T. G., Hirschwerk D. A., Kim E. J., Kozel Z. M., Marrast L. M., Mogavero J. N., Osorio G. A., Qiu M., Zanos T. P., Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York city area. JAMA 323, 2052–2059 (2020). - PMC - PubMed
- Seow J., Graham C., Merrick B., Acors S., Pickering S., Steel K. J. A., Hemmings O., O’Byrne A., Kouphou N., Galao R. P., Betancor G., Wilson H. D., Signell A. W., Winstone H., Kerridge C., Huettner I., Jimenez-Guardeño J. M., Lista M. J., Temperton N., Snell L. B., Bisnauthsing K., Moore A., Green A., Martinez L., Stokes B., Honey J., Izquierdo-Barras A., Arbane G., Patel A., Tan M. K. I., O’Connell L., O’Hara G., MacMahon E., Douthwaite S., Nebbia G., Batra R., Martinez-Nunez R., Shankar-Hari M., Edgeworth J. D., Neil S. J. D., Malim M. H., Doores K. J., Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat. Microbiol. 5, 1598–1607 (2020). - PMC - PubMed
- Long Q.-X., Tang X.-J., Shi Q.-L., Li Q., Deng H.-J., Yuan J., Hu J.-L., Xu W., Zhang Y., Lv F.-J., Su K., Zhang F., Gong J., Wu B., Liu X.-M., Li J.-J., Qiu J.-F., Chen J., Huang A.-L., Clinical and immunological assessment of asymptomatic SARS-CoV-2 infections. Nat. Med. 26, 1200–1204 (2020). - PubMed
Grants and funding
- R01 AI158013/AI/NIAID NIH HHS/United States
- R21 AI167648/AI/NIAID NIH HHS/United States
- K99 EB030587/EB/NIBIB NIH HHS/United States
- R35 GM138361/GM/NIGMS NIH HHS/United States
- R01 AI141003/AI/NIAID NIH HHS/United States
- R01 HL105704/HL/NHLBI NIH HHS/United States
- K24 AI144048/AI/NIAID NIH HHS/United States
- R01 GM130900/GM/NIGMS NIH HHS/United States
- T32 AI060530/AI/NIAID NIH HHS/United States
- U24 GM132013/GM/NIGMS NIH HHS/United States