CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2 - PubMed (original) (raw)
. 2021 Jul 28;7(7):1156-1165.
doi: 10.1021/acscentsci.0c01537. Epub 2021 Jun 30.
Wenqing Yin 2, Marc A Napoleon 2, Ellen L Suder 3 4, Jacob Berrigan 3, Qing Zhao 1, Judith Olejnik 3 4, Kevin Brown Chandler 5, Chaoshuang Xia 5, Jared Feldman 6, Blake M Hauser 6, Timothy M Caradonna 6, Aaron G Schmidt 6 7, Suryaram Gummuluru 3, Elke Mühlberger 3 4, Vipul Chitalia 2, Catherine E Costello 5, Nader Rahimi 1
Affiliations
- PMID: 34341769
- PMCID: PMC8265543
- DOI: 10.1021/acscentsci.0c01537
CD209L/L-SIGN and CD209/DC-SIGN Act as Receptors for SARS-CoV-2
Razie Amraei et al. ACS Cent Sci. 2021.
Abstract
As the COVID-19 pandemic continues to spread, investigating the processes underlying the interactions between SARS-CoV-2 and its hosts is of high importance. Here, we report the identification of CD209L/L-SIGN and the related protein CD209/DC-SIGN as receptors capable of mediating SARS-CoV-2 entry into human cells. Immunofluorescence staining of human tissues revealed prominent expression of CD209L in the lung and kidney epithelia and endothelia. Multiple biochemical assays using a purified recombinant SARS-CoV-2 spike receptor-binding domain (S-RBD) or S1 encompassing both N termal domain and RBD and ectopically expressed CD209L and CD209 revealed that CD209L and CD209 interact with S-RBD. CD209L contains two _N_-glycosylation sequons, at sites N92 and N361, but we determined that only site N92 is occupied. Removal of the _N_-glycosylation at this site enhances the binding of S-RBD with CD209L. CD209L also interacts with ACE2, suggesting a role for heterodimerization of CD209L and ACE2 in SARS-CoV-2 entry and infection in cell types where both are present. Furthermore, we demonstrate that human endothelial cells are permissive to SARS-CoV-2 infection, and interference with CD209L activity by a knockdown strategy or with soluble CD209L inhibits virus entry. Our observations demonstrate that CD209L and CD209 serve as alternative receptors for SARS-CoV-2 in disease-relevant cell types, including the vascular system. This property is particularly important in tissues where ACE2 has low expression or is absent and may have implications for antiviral drug development.
© 2021 The Authors. Published by American Chemical Society.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Figure 1
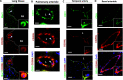
CD209L is expressed in lung as well as renal epithelial and endothelial cells: PFA fixed human lung, renal, and temporal arteriole tissues were subjected to immunofluorescence staining. Lung tissue stained with anti-MUC1, anti-CD31, and anti-CD209L antibodies. (A) Type II alveoli epithelial cells of alveoli were positive for CD209L (red) and MUC1 (green). (B) Pulmonary arteriole endothelial cells were positive for CD31 (green) and CD209L (red). (C) Endothelial cells of the temporal arteriole were stained with CD209L (green) and CD31 (red). (D) Renal endothelial cells were positive for CD209L (red) and CD31 (green). White arrowhead points to the alveolar cell, AS = alveolar space, and white dotted line corresponds to alveolar septa. Image magnification, white bars = 50 μm.
Figure 2
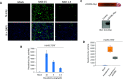
Endothelial cells are permissive to SARS-CoV-2 virus, and soluble CD209L neutralizes viral entry. (A) HUVEC-TERT cells seeded in chamber slides were mock-infected or infected with SARS-CoV-2 at the indicated MOIs. Cells were fixed at 1 day post infection and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue). Image magnification 50 μm. (B) HUVEC-TERT cells (2 × 104/well, 96-well plate, and quadruple/group) were infected with pseudovirus with different concentrations. After 24 h, cells were processed and subjected to a luciferase activity assay, and representative data are shown. (C) Schematic of Myc-tagged soluble CD209 (sCD209L-Myc) and Western blot analysis of sCD209L-Myc. (D) HUVEC-TERT cells (2 × 104/well, 96-well plate, and triplicate/group) were infected with mock, pseudotyped virus (10 ng/mL) with control conditioned medium (CM) or CM containing sCD209L (approximate concentration 1.5 μg/mL). After 24 h, cells were analyzed for luciferase activity. P < 0.05.
Figure 3
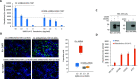
CD209L mediates SARS-CoV-2 entry and infection in endothelial cells. (A) HUVEC-TERT cells expressing control shRNA or CD209L-shRNA (2 × 104/well, 96-well plate, and triplicate/group) were infected with different amounts of SARS-CoV-2 pseudotyped lentivirus. After 24 h, cells were processed and subjected to luciferase activity, and representative data are shown. (B) HUVEC-TERT cells expressing control shRNA or CD209L-shRNA seeded in chamber slides (triplicate/group) were infected with SARS-CoV-2 at the indicated MOIs. Cells were fixed at 1 day post infection and stained with an antibody directed against the viral nucleoprotein, N (green). Cell nuclei were stained with DAPI (blue). Image magnification 50 μm. Quantification of N protein positive cells is shown. P < 0.05. (C) Western blot analysis showing expression of CD209L, CD209, and ACE2. (D) HEK-293 cells expressing CD209L, CD209, or ACE2 (2 × 104/well, 96-well plate, and quadruple wells/group) were infected with pseudotyped virus (10 ng/mL) or mock virus. After 24 h, cells were analyzed for luciferase activity. P < 0.05.
Figure 4
CD209L and CD209 bind to SARS-CoV-2-S-RBD. (A) Schematic of Fc-CoV-2-S-RBD; expression of Fc-CoV-2-S-RBD and CD209L in HEK-293 is shown. (B) Immunoprecipitation assay demonstrates the binding of CD209L with Fc-CoV-2-S-RBD. (C) Schematic and Coomassie blue stain of CoV-2-S-RBD-HIS are shown. (D) Far-Western blot analysis shows the binding of HIS-STRP-tagged CoV-2-S-RBD with CD209L. (E) Western blot analysis of CD209L, CD209, and Fc-ACE2 ectopically expressed in HEK-293 cells. (F) CoV-2-S-RBD-HIS applied onto a PFVD membrane with varying concentrations as indicated in the figure legend via a dot blot apparatus. The membranes, after blocked with 5% BSA, were incubated with cell lysates derived from HEK-293 cells expressing Fc-ACE-2-Myc, CD209L-Myc, or CD209-Myc, and the binding of ACE2, CD209L, and CD209 to CoV-2-S-RBD was detected with anti-Myc antibody. Quantification of the dot blots is shown. AU, arbitrary unit. ImageJ software was used to quantify the dot blots.
Figure 5
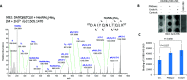
CD209L is _N_-glycosylated, and _N_-glycosylation modulates its interaction with SARS-CoV2 spike protein. (A) HCD MS/MS spectrum of m/z 1505.1470, assigned as the [M + 2H]2+ of the glycopeptide 87DAIYQNLTQLK97 + HexNAc2Hex8, corresponding to CD209L site N92 bearing a Man8 glycan. Stepped-collision energy (15/25/35% NCE) was used. The fragment ions do not retain the glycan moiety. p = peptide, N = N-acetylhexosamine (HexNAc), H = hexose (Hex). (B) Dot blot of CD209L/HEK-293 cell lysate under control conditions (no treatment) or treated with PNGase F or Endo H. Dot blot of HEK-293 cell lysate was used as a negative control. Following treatment of lysate, the spike protein receptor-binding domain (S-RBD-HIS) was incubated with the immobilized lysate, followed by detection with anti-HIS antibody. (C) Quantification of blots is shown. AU, arbitrary unit. ImageJ software was used to quantify the dot blots.
Update of
- CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2.
Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, Olejnik J, Chandler KB, Xia C, Feldman J, Hauser BM, Caradonna TM, Schmidt AG, Gummuluru S, Muhlberger E, Chitalia V, Costello CE, Rahimi N. Amraei R, et al. bioRxiv [Preprint]. 2021 Jun 14:2020.06.22.165803. doi: 10.1101/2020.06.22.165803. bioRxiv. 2021. PMID: 32607506 Free PMC article. Updated. Preprint.
Similar articles
- CD209L/L-SIGN and CD209/DC-SIGN act as receptors for SARS-CoV-2.
Amraei R, Yin W, Napoleon MA, Suder EL, Berrigan J, Zhao Q, Olejnik J, Chandler KB, Xia C, Feldman J, Hauser BM, Caradonna TM, Schmidt AG, Gummuluru S, Muhlberger E, Chitalia V, Costello CE, Rahimi N. Amraei R, et al. bioRxiv [Preprint]. 2021 Jun 14:2020.06.22.165803. doi: 10.1101/2020.06.22.165803. bioRxiv. 2021. PMID: 32607506 Free PMC article. Updated. Preprint. - C-type Lectin CD209L/L-SIGN and CD209/DC-SIGN: Cell Adhesion Molecules Turned to Pathogen Recognition Receptors.
Rahimi N. Rahimi N. Biology (Basel). 2020 Dec 22;10(1):1. doi: 10.3390/biology10010001. Biology (Basel). 2020. PMID: 33375175 Free PMC article. Review. - CD209L (L-SIGN) is a receptor for severe acute respiratory syndrome coronavirus.
Jeffers SA, Tusell SM, Gillim-Ross L, Hemmila EM, Achenbach JE, Babcock GJ, Thomas WD Jr, Thackray LB, Young MD, Mason RJ, Ambrosino DM, Wentworth DE, Demartini JC, Holmes KV. Jeffers SA, et al. Proc Natl Acad Sci U S A. 2004 Nov 2;101(44):15748-53. doi: 10.1073/pnas.0403812101. Epub 2004 Oct 20. Proc Natl Acad Sci U S A. 2004. PMID: 15496474 Free PMC article. - ACE2-Independent Alternative Receptors for SARS-CoV-2.
Lim S, Zhang M, Chang TL. Lim S, et al. Viruses. 2022 Nov 16;14(11):2535. doi: 10.3390/v14112535. Viruses. 2022. PMID: 36423144 Free PMC article. Review. - SARS-CoV-2 Spike Protein Interacts with Multiple Innate Immune Receptors.
Gao C, Zeng J, Jia N, Stavenhagen K, Matsumoto Y, Zhang H, Li J, Hume AJ, Mühlberger E, van Die I, Kwan J, Tantisira K, Emili A, Cummings RD. Gao C, et al. bioRxiv [Preprint]. 2020 Jul 30:2020.07.29.227462. doi: 10.1101/2020.07.29.227462. bioRxiv. 2020. PMID: 32766577 Free PMC article. Preprint.
Cited by
- Early Steps of Individual Multireceptor Viral Interactions Dissected by High-Density, Multicolor Quantum Dot Mapping in Living Cells.
Mateos N, Gutierrez-Martinez E, Angulo-Capel J, Carlon-Andres I, Padilla-Parra S, Garcia-Parajo MF, Torreno-Pina JA. Mateos N, et al. ACS Nano. 2024 Oct 22;18(42):28881-28893. doi: 10.1021/acsnano.4c09085. Epub 2024 Oct 10. ACS Nano. 2024. PMID: 39387532 Free PMC article. - Clinical glycoproteomics: methods and diseases.
Wang Y, Lei K, Zhao L, Zhang Y. Wang Y, et al. MedComm (2020). 2024 Oct 4;5(10):e760. doi: 10.1002/mco2.760. eCollection 2024 Oct. MedComm (2020). 2024. PMID: 39372389 Free PMC article. Review. - Interactions of SARS-CoV-2 with Human Target Cells-A Metabolic View.
Eisenreich W, Leberfing J, Rudel T, Heesemann J, Goebel W. Eisenreich W, et al. Int J Mol Sci. 2024 Sep 16;25(18):9977. doi: 10.3390/ijms25189977. Int J Mol Sci. 2024. PMID: 39337465 Free PMC article. Review. - The Functions of SARS-CoV-2 Receptors in Diabetes-Related Severe COVID-19.
Drzymała A. Drzymała A. Int J Mol Sci. 2024 Sep 5;25(17):9635. doi: 10.3390/ijms25179635. Int J Mol Sci. 2024. PMID: 39273582 Free PMC article. Review. - Unprecedented selectivity for homologous lectin targets: differential targeting of the viral receptors L-SIGN and DC-SIGN.
Delaunay C, Pollastri S, Thépaut M, Cavazzoli G, Belvisi L, Bouchikri C, Labiod N, Lasala F, Gimeno A, Franconetti A, Jiménez-Barbero J, Ardá A, Delgado R, Bernardi A, Fieschi F. Delaunay C, et al. Chem Sci. 2024 Aug 27;15(37):15352-66. doi: 10.1039/d4sc02980a. Online ahead of print. Chem Sci. 2024. PMID: 39246372 Free PMC article.
References
- Bhatraju P. K.; Ghassemieh B. J.; Nichols M.; Kim R.; Jerome K. R.; Nalla A. K.; Greninger A. L.; Pipavath S.; Wurfel M. M.; Evans L.; Kritek P. A.; West T. E.; Luks A.; Gerbino A.; Dale C. R.; Goldman J. D.; O’Mahony S.; Mikacenic C. Covid-19 in Critically Ill Patients in the Seattle Region-Case Series. N. Engl. J. Med. 2020, 382, 2012.10.1056/NEJMoa2004500. - DOI - PMC - PubMed
- Huang C.; Wang Y.; Li X.; Ren L.; Zhao J.; Hu Y.; Zhang L.; Fan G.; Xu J.; Gu X.; Cheng Z.; Yu T.; Xia J.; Wei Y.; Wu W.; Xie X.; Yin W.; Li H.; Liu M.; Xiao Y.; Gao H.; Guo L.; Xie J.; Wang G.; Jiang R.; Gao Z.; Jin Q.; Wang J.; Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395 (10223), 497–506. 10.1016/S0140-6736(20)30183-5. - DOI - PMC - PubMed
Grants and funding
- S10 OD021728/OD/NIH HHS/United States
- UC7 AI095321/AI/NIAID NIH HHS/United States
- R21 CA193958/CA/NCI NIH HHS/United States
- T32 HL007035/HL/NHLBI NIH HHS/United States
- T32 GM007753/GM/NIGMS NIH HHS/United States
- R24 GM134210/GM/NIGMS NIH HHS/United States
- R01 AI146779/AI/NIAID NIH HHS/United States
- T32 GM008313/GM/NIGMS NIH HHS/United States
- R01 AI064099/AI/NIAID NIH HHS/United States
- R01 HL132325/HL/NHLBI NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
- T32 AI007245/AI/NIAID NIH HHS/United States
- R01 AG060890/AG/NIA NIH HHS/United States
- S10 OD010724/OD/NIH HHS/United States
- R21 CA191970/CA/NCI NIH HHS/United States
- R01 AI133486/AI/NIAID NIH HHS/United States
- P30 AI042853/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous