Identification of an Intestinal Microbiota Signature Associated With the Severity of Necrotic Enteritis - PubMed (original) (raw)
Identification of an Intestinal Microbiota Signature Associated With the Severity of Necrotic Enteritis
Qing Yang et al. Front Microbiol. 2021.
Abstract
Necrotic enteritis (NE), an economically devastating disease of poultry caused by pathogenic Clostridium perfringens, is known to induce small intestinal lesions and dysbiosis. However, the intestinal microbes that are associated with NE severity are yet to be characterized. Here, we investigated the link between the ileal microbiota and disease severity in a chicken model of clinical NE using 16S rRNA gene sequencing. Our results indicated that richness and Shannon Index of the ileal microbiota were drastically reduced (p<0.01) as NE was exacerbated. While the relative abundance of C. perfringens increased from 0.02% in healthy chickens to 58-70% in chickens with severe infection, a majority of the ileal microbes were markedly diminished, albeit varying in their sensitivity to NE. Compositionally, a large group of ileal microbes showed a significant correlation with NE severity. Firmicutes, such as group A and B Lactobacillus, Lactobacillus reuteri, Subdoligranulum variabile, Mediterraneibacter, and Staphylococcus as well as two genera of Actinobacteria (Corynebacterium and Kocuria) and two highly related Cyanobacteria were progressively declined as NE was aggravated. Other Firmicutes, such as Weissella, Romboutsia, Kurthia, Cuneatibacter, Blautia, and Aerococcus, appeared much more sensitive and were rapidly abolished in chickens even with mild NE. On the other hand, Enterococcus cecorum and two Escherichia/Shigella species were only enriched in the ileal microbiota of chickens with extremely severe NE, while several other species such as Streptococcus gallolyticus and Bacteroides fragilis remained unaltered by NE. Functionally, secondary bile acid biosynthesis was predicted to be suppressed by NE, while biosynthesis of aromatic and branched-amino acids and metabolism of a majority of amino acids were predicted to be enhanced in the ileum of NE-afflicted chickens. These intestinal microbes showing a strong correlation with NE severity may provide important leads for the development of novel diagnostic or therapeutic approaches to NE and possibly other enteric diseases.
Keywords: 16S rRNA gene sequencing; Clostridium perfringens; dysbiosis; microbiome; microbiota; necrotic enteritis; poultry.
Copyright © 2021 Yang, Liu, Wang, Robinson, Whitmore, Stewart, Zhao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Figure 1
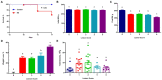
Survival rate, growth performance, and total bacterial population among chickens with varying severities of necrotic enteritis (NE). A group of 90 male Cobb broilers were infected sequentially with Eimeria maxima on day 10 and Clostridium perfringens on day 14, while another 10 chickens were mock-infected as negative controls. Animals were separated into five groups according to their respective intestinal lesion scores measured on day 17. Animal survival was recorded daily between days 14 and 17 (A) and was compared between infected and mock-infected healthy animals using the log-rank test. Body weights of individual animals were recorded on day 10 (B) and day 17 (C). Weight loss (%) of infected chickens during days 10–17 (D) was calculated relative to healthy controls. Total bacteria number in the ileal digesta (E) was also estimated using qPCR. All data were expressed as means±SEM. Different superscripts denote significance (p<0.05) based on one-way ANOVA and Tukey’s post-hoc test.
Figure 2
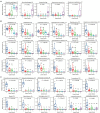
Diversity and composition of the ileal microbiota among chickens with varying severities of NE. A group of 90 male Cobb broilers were subjected to NE, while another 10 chickens in the control group were mock-infected. Animals were divided into five groups based on lesion scores. The number of observed amplicon sequence variants (ASVs; A) and Shannon index (B) of the ileal microbiome were estimated and visualized using box and whisker plots. Each box indicates median, 25 and 75th percentiles, while whiskers extend to 1.5 interquartile range. Significance was measured using the Kruskal-Wallis test, and pairwise comparisons were implemented using the Wilcoxon rank sum test. Different superscripts denote significance (p<0.05) in pairwise comparisons. (C) Principal coordinates analysis (PCoA) plots of weighted UniFrac distances. Significance was determined using permutational multivariate ANOVA (PERMANOVA). Relative abundances of top five phyla (D), top five families (E), top five genera (F), and top 10 ASVs (G) in the ileal microbiota were shown.
Figure 3
Differential enrichment of the ileal bacteria between chickens with mild and severe NE. After induction of NE, the ileal digesta of broilers were collected and subjected to 16S rRNA gene sequencing. Intestinal lesions of individual animals were scored. Linear discriminant analysis (LDA) effect size (LEfSe) analysis was performed to identify differentially abundant bacteria at the phylum (A), order (B), family (C), genus (D), and ASV levels (E) between score-1 mild and score-6 severe NE chickens with cut-offs p<0.05 and LDA score>2.0. (F) Phylogenetic relationships of differentially enriched bacteria. Differentially enriched bacterial taxa were represented by rings with the phylum shown in the outermost ring and the species in the innermost ring.
Figure 4
Correlation between relative abundance of the ileal bacteria and the severity of NE. The Spearman correlation analysis was performed between the ileal microbiota profile and the intestinal lesion score and weight loss during days 10–17. (A) Spearman correlation at the family and genus levels. (B) Heatmap showing log2 transformations of the fold changes of the ileal bacteria among different groups of NE at the family and genus levels relative to score-0 healthy controls. (C) Spearman correlation at the ASV level. Significance of Spearman correlation analysis was corrected with the Benjamini-Hochberg procedure. *FDR<0.05, **FDR<0.01.
Figure 5
Relative abundances of representative ileal bacteria showing a significant correlation with the severity of NE. Ileal bacteria were significantly enriched (A), progressively declined (B), or abruptly abolished (C) in response to NE. In the box and whisker plots, each box indicates median, 25 and 75th percentiles, while whiskers extend to 1.5 interquartile range. Significance was measured using the Kruskal-Wallis test and indicated on the top of each plot. Pairwise comparisons were further implemented using the Wilcoxon rank sum test, and the significance (p<0.05) was denoted by different superscripts.
Figure 6
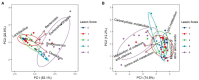
Biplots of principal component analysis showing relative contributions of top five genera (A) and top five predicted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (B) to the divergence of the ileal microbiota in response to NE. Each biplot line shows the direction of the change, with the length of each line indicating the degree of correlation with ordination axes.
Figure 7
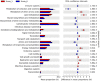
Differential enrichment of predicted KEGG pathways between mild and severe NE. Functional potentials of ileal microbiota of score-1 and score-6 chickens were predicted using PICRUSt2 and level-2 KEGG pathways were compared in Statistical Analysis of Metagenomic Profiles (STAMP). Each bar represents a mean proportion (%) and SEM of a predicted KEGG pathway. The 95% CIs of the differences in mean proportions between score-1 and score-6 chickens were also shown. Significance was determined by two-sided White’s non-parametric _t_-test, corrected with the Benjamini-Hochberg procedure, and indicated as q values on the right. Only those pathways showing q<0.05 were displayed.
Figure 8
Classification of the ileal bacteria and their taxonomies based on their sensitivity to NE. While a majority of bacteria were abruptly vanished (highly sensitive) or gradually diminished (moderately sensitive) by NE, a few bacteria were only enriched during severe disease (minimally sensitive) or remained unchanged (resistant) by NE. Reduced bacterial taxa were indicated in green, and enriched bacteria were highlighted in red, while unaltered bacteria were indicated in bold black.
Similar articles
- Perturbations of the ileal mycobiota by necrotic enteritis in broiler chickens.
Yang Q, Liu J, Robinson KJ, Whitmore MA, Stewart SN, Zhang G. Yang Q, et al. J Anim Sci Biotechnol. 2021 Oct 9;12(1):107. doi: 10.1186/s40104-021-00628-5. J Anim Sci Biotechnol. 2021. PMID: 34625122 Free PMC article. - Analysis of gut microbiota and the effect of lauric acid against necrotic enteritis in Clostridium perfringens and Eimeria side-by-side challenge model.
Yang WY, Lee Y, Lu H, Chou CH, Wang C. Yang WY, et al. PLoS One. 2019 May 31;14(5):e0205784. doi: 10.1371/journal.pone.0205784. eCollection 2019. PLoS One. 2019. PMID: 31150394 Free PMC article. - Microbial metabolite deoxycholic acid controls Clostridium perfringens-induced chicken necrotic enteritis through attenuating inflammatory cyclooxygenase signaling.
Wang H, Latorre JD, Bansal M, Abraha M, Al-Rubaye B, Tellez-Isaias G, Hargis B, Sun X. Wang H, et al. Sci Rep. 2019 Oct 10;9(1):14541. doi: 10.1038/s41598-019-51104-0. Sci Rep. 2019. PMID: 31601882 Free PMC article. - Microbial shifts associated with necrotic enteritis.
Antonissen G, Eeckhaut V, Van Driessche K, Onrust L, Haesebrouck F, Ducatelle R, Moore RJ, Van Immerseel F. Antonissen G, et al. Avian Pathol. 2016 Jun;45(3):308-12. doi: 10.1080/03079457.2016.1152625. Avian Pathol. 2016. PMID: 26950294 Review. - Poultry management: a useful tool for the control of necrotic enteritis in poultry.
Tsiouris V. Tsiouris V. Avian Pathol. 2016 Jun;45(3):323-5. doi: 10.1080/03079457.2016.1154502. Avian Pathol. 2016. PMID: 26950433 Review.
Cited by
- Comparison of gut microbiota immunity and pathology in specific-pathogen-free chickens with glandular and muscular gastritis using different methods.
Yan Z, Yang S, Lin S, Zhao Z, Liu Y, Yin B, Yi Y, Song S, Zhang R, Huang Z. Yan Z, et al. Front Vet Sci. 2024 Jun 3;11:1343768. doi: 10.3389/fvets.2024.1343768. eCollection 2024. Front Vet Sci. 2024. PMID: 38887537 Free PMC article. - The effect of supplemental arginine on the gut microbial homeostasis of broilers during sub-clinical necrotic enteritis challenge.
Fathima S, Al Hakeem WG, Shanmugasundaram R, Lourenco J, Selvaraj RK. Fathima S, et al. Front Physiol. 2024 Sep 17;15:1463420. doi: 10.3389/fphys.2024.1463420. eCollection 2024. Front Physiol. 2024. PMID: 39355151 Free PMC article. - Effects of challenge with Clostridium perfringens, Eimeria and both on ileal microbiota of yellow feather broilers.
Feng X, Li T, Zhu H, Liu L, Bi S, Chen X, Zhang H. Feng X, et al. Front Microbiol. 2022 Dec 1;13:1063578. doi: 10.3389/fmicb.2022.1063578. eCollection 2022. Front Microbiol. 2022. PMID: 36532499 Free PMC article. - Lactiplantibacillus plantarum ST-III-fermented milk improves autistic-like behaviors in valproic acid-induced autism spectrum disorder mice by altering gut microbiota.
Zhang Y, Guo M, Zhang H, Wang Y, Li R, Liu Z, Zheng H, You C. Zhang Y, et al. Front Nutr. 2022 Nov 24;9:1005308. doi: 10.3389/fnut.2022.1005308. eCollection 2022. Front Nutr. 2022. PMID: 36505260 Free PMC article. - Correlation between gut microbiota and the development of Graves' disease: A prospective study.
Deng Y, Wang J, Xie G, Zou G, Li S, Zhang J, Cai W, Xu J. Deng Y, et al. iScience. 2023 Jun 28;26(7):107188. doi: 10.1016/j.isci.2023.107188. eCollection 2023 Jul 21. iScience. 2023. PMID: 37485373 Free PMC article.
References
- Altermann E., Russell W. M., Azcarate-Peril M. A., Barrangou R., Buck B. L., Mcauliffe O., et al. . (2005). Complete genome sequence of the probiotic lactic acid bacterium lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. U. S. A. 102, 3906–3912. 10.1073/pnas.0409188102, PMID: - DOI - PMC - PubMed
- Anderson M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral. Ecol. 26, 32–46. 10.1111/j.1442-9993.2001.01070.pp.x - DOI
LinkOut - more resources
Full Text Sources