Microtubule-based transport is essential to distribute RNA and nascent protein in skeletal muscle - PubMed (original) (raw)
Microtubule-based transport is essential to distribute RNA and nascent protein in skeletal muscle
Lance T Denes et al. Nat Commun. 2021.
Abstract
While the importance of RNA localization in highly differentiated cells is well appreciated, basic principles of RNA localization in skeletal muscle remain poorly characterized. Here, we develop a method to detect and quantify single molecule RNA localization patterns in skeletal myofibers, and uncover a critical role for directed transport of RNPs in muscle. We find that RNAs localize and are translated along sarcomere Z-disks, dispersing tens of microns from progenitor nuclei, regardless of encoded protein function. We find that directed transport along the lattice-like microtubule network of myofibers becomes essential to achieve this localization pattern as muscle development progresses; disruption of this network leads to extreme accumulation of RNPs and nascent protein around myonuclei. Our observations suggest that global active RNP transport may be required to distribute RNAs in highly differentiated cells and reveal fundamental mechanisms of gene regulation, with consequences for myopathies caused by perturbations to RNPs or microtubules.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
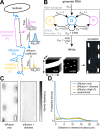
Fig. 1. RNAs can be reproducibly and accurately detected in myofibers by HCR FISH and are dispersed in the myofiber cytoplasm.
A Schematic describing experimental strategy to label RNAs and proteins of interest in adult skeletal muscle. B Transcript length (nucleotides, nt) and abundance in tibialis anterior muscle (transcripts per million, TPM) for each RNA studied; colors represent encoded protein localization. C Representative FISH images for each RNA studied. Scale bars: 5 μm. D RNA density (spots/µm3) measured from FISH images compared with transcripts per million (TPM) values from a tibialis anterior RNAseq dataset. Dotted line is lower limit of detection (LLOD). Trendline: LLS regression; Pearson R = 0.98; p < 0.05, Wald test. E RNA densities compared across separate experiments. Black bars are the mean ± s.d. of RNA density. Experiment 1: n = 10 myofibers (Polr2a, Hist1h1c, Ttn, GFP) or n = 9 myofibers (Vcl, Dmd, Hnrnpa2b1, Myom1, Gapdh). Experiment 2: n = 3 myofibers. Trendline: LLS regression; Pearson R = 0.97; p < 0.05, Wald test. F Schematic describing percent dispersion calculation with example cumulative distribution function (CDF) for Polr2a RNA. G Percent dispersion for each RNA studied, points are colored as in Fig. 1B. Dotted line indicates fully uniform dispersion. H Mean Gapdh FISH signal intensity of myofiber in C plotted for 40 µm along the longitudinal axis (top). The power spectral density of this signal (bottom).
Fig. 2. RNAs co-localize with Z-disks and microtubules.
A Representative image of IF/FISH co-labeling of Ttn RNAs (red), tubulin protein (microtubules, green), and telethonin protein (Z-disks, blue) in an isolated myofiber. Scale bar: 5 µm. B Zoomed-in regions of IF/FISH co-labeled myofibers as in A for each RNA studied. Scale bars: 1 µm. C Schematic describing the computational pipeline used to assess mRNA proximity to cytoskeleton from images of IF/FISH co-labeled myofibers (A). D Distances from cytoplasmic spots (blue boxes) to Z-disks (left) and microtubules (right) compared to null distributions generated from randomly selected cytoplasmic coordinates (gray boxes). Data from n = 10 myofibers (Polr2a, Hist1h1c, Ttn, GFP) or n = 9 myofibers (Vcl, Dmd, Hnrnpa2b1, Myom1, Gapdh). Spots located within 2 pixels (~0.1 μm) of either filament were considered “cytoskeleton co-localized” (purple shaded region, percentages). Box plots show minimum, first quartile, median, third quartile, and maximum. *p < 0.03, **p < 10−4, Two-sided Mann–Whitney U test. E Distances from cytoskeleton-associated spots to ZMIs (blue boxes) compared to a null distribution generated from randomly selected coordinates along the cytoskeleton (gray boxes). Number of myofibers same as D. Box plots show minimum, first quartile, median, third quartile, and maximum. *p < 0.03, **p < 10−4, Two-sided Mann–Whitney U test.
Fig. 3. Chrne RNA produced in postsynaptic nuclei is confined to the NMJ region.
A Representative image of FISH for Ttn (green) and Chrne (red) RNA in an extrasynaptic region of an isolated EDL myofiber. Scale bars: 2 µm. B Representative image of FISH for Ttn (green) and Chrne (red) RNA in the NMJ of an isolated EDL myofiber. Scale bars: 2 µm. C Quantification of RNA density for Ttn and Chrne RNAs in extrasynaptic (blue) and NMJ regions (red) of myofibers. Extrasynaptic regions: n = 5 myofibers; NMJ regions: n = 8 myofibers. *p < 0.05, **p < 10−4. Two-sided Mann–Whitney U test. Dotted line indicates lower limit of detection (LLOD). D Tubulin IF in NMJ region of an isolated myofiber. Scale bars: 2 µm. E IF/FISH co-labeling of Ttn RNA (red), tubulin protein (microtubules, green), and telethonin protein (Z-disks, blue) at the NMJ of an isolated myofiber. Scale bar: 2 µm. F IF/FISH co-labeling of Chrne RNA (red), tubulin protein (microtubules, green), and telethonin protein (Z-disks, blue) at the NMJ of an isolated myofiber. Scale bar: 2 µm. G Large field of view image of IF/FISH co-labeling of Chrne RNA (red), tubulin protein (microtubules, green), and telethonin protein (Z-disks, blue). * indicates Chrne RNAs in the extrasynaptic myofiber neighboring the NMJ. † indicates extrasynaptic nuclei containing Chrne mRNAs. ^ indicates NMJ region containing high concentration of Chrne RNA. Scale bar: 10 µm.
Fig. 4. RNAs accumulate in the perinuclear region of myofibers after microtubule ablation.
A Schematic describing microtubule depolymerization experiment. B IF of tubulin (green) and telethonin (red) protein in myofibers from A. Scale bars: 5 µm. C Representative FISH images of Ttn RNA in myofibers from A. Scale bars: 5 µm. D Same as C but Polr2a RNA. Scale bars: 5 µm. E Intranuclear position relative to centroid (C) and periphery (P) for intranuclear spots and distance to nearest nucleus for cytoplasmic spots; shown for Ttn RNA. Bar colors correspond to treatment condition and shaded area denotes perinuclear region, defined as <2 μm from nuclear periphery (n.p.). Number of myofibers per RNA and condition are Dmd: DMSO-8, nocodazole-8, washout-6; Gapdh: DMSO-9, nocodazole-8, washout-3; Hist1h1c: DMSO-7, nocodazole-12, washout-4; Hnrnpa2b1: DMSO-8, nocodazole-5, washout-5; Myom1: DMSO-5, nocodazole-5, washout-5; Polr2a: DMSO-5, nocodazole-5, washout-5; Ttn: DMSO-11, nocodazole-8, washout-5; Vcl: DMSO-5, nocodazole-5, washout-5. F Same as E but Polr2a RNA. G Fraction of total spots in the perinuclear region for each gene. Colors as in E. Bars are mean ± SEM. Black dots are individual images. Number of myofibers as in E. *p < 0.05, Two-sided Mann–Whitney U test. H Representative images for each RNA showing zoomed-in regions of individual nuclei. Scale bars: 1 µm. I Cytoplasmic and perinuclear blob intensities for Gapdh RNA in each experimental condition. Colors as in E. J Intensity of perinuclear blobs. Bars are 95th percentile ±95% CI. n.s. not significant, *p < 0.05, by confidence interval overlap. Confidence intervals were estimated by bootstrapping. K Same as J but for cytoplasmic blobs. Number of myofibers as in E. L Schematic describing the effects of Actinomycin D and nocodazole treatments on cytoplasmic RNA concentration. M For each RNA, the fraction remaining in the cytoplasm after 18 h nocodazole treatment compared with the fraction predicted by our decay model. Trend line: LLS regression, slope = 0.84; pearson R = 0.66; p < 0.05, Wald test.
Fig. 5. Muscle development restricts microtubule-independent mRNA dispersion.
A Polr2a FISH in C2C12 myoblast after 6 h control (DMSO) culture. Scale bars: 2 µm. B Polr2a FISH in C2C12 myoblast after 6 h culture with nocodazole. Scale bars: 2 µm. C Polr2a FISH in C2C12 myotube after 6 h control (DMSO) culture. Scale bars: 2 µm. D Polr2a FISH in C2C12 myotube after 6 h culture with nocodazole. Scale bars: 2 µm. E Polr2a FISH in myofiber after 6 h control (DMSO) culture. Scale bars: 2 µm. F Polr2a FISH in myofiber after 6 hr culture with nocodazole. Scale bars: 2 µm. G Percent dispersion for Polr2a RNA in C2C12 myoblasts, C2C12 myotubes, and isolated myofibers treated with nocodazole or control (DMSO) for 6 h. Black lines are medians. n = 5 myotubes and myofibers per condition. n = 10 and 12 cells for nocodazole and DMSO-treated myoblasts, respectively. *p < 0.05, **p < 0.01, Two-sided Mann–Whitney U test.
Fig. 6. Nocodazole treatment causes accumulation of ribosomes and nascent protein around myonuclei.
A Ribosomes were detected in isolated myofibers using HCR FISH for 28S ribosomal RNA. Representative images of IF/FISH co-labeling for tubulin (green) and telethonin (blue) proteins with rRNA (red) in myofibers. Scale bars: 5 µm. B Active translation was detected in isolated myofibers using in situ puromycylation. Representative images of puromycin detection (red) with tubulin (green) and telethonin (blue) IF in myofibers. Scale bars: 5 µm. C Live myofibers were isolated and cultured with nocodazole or DMSO (control) for 18 or 36 hr. At each time point, rRNA FISH was performed. D Representative images of rRNA FISH signal in myofibers from C. Scale bars: 5 µm. E Perinuclear to cytoplasmic relative rRNA signal density (pixel intensity per µm3) in myofibers from C. Mean ± 95% CI. n = 5 myofibers per condition. *p < 0.05, Two-sided Mann–Whitney U test. F Live myofibers were isolated and cultured with nocodazole or DMSO (control) for 18 or 36 h. At each time point, in situ puromycylation was performed as described in B. G Representative images of puromycin IF in myofibers from F. Scale bars: 5 µm. H Perinuclear to cytoplasmic relative puromycin signal density in myofibers from F. Mean ± 95% CI. n = 5 myofibers per condition. *p < 0.05, Two-sided Mann–Whitney U test.
Fig. 7. Regulators of RNP granule formation, transport, and translation co-accumulate at the nuclear periphery of nocodazole-treated myofibers.
A Representative images of Co-IF against the nuclear pore complex (NPC, labeled with mAb 414) and Kif1C in myofibers cultured for 18 h in the presence of nocodazole or control (DMSO). Scale bars: 2 µm. B Representative images of Co-IF against G3bp1 and Fxr1p in myofibers cultured for 18 h in the presence of nocodazole or control (DMSO). See also Supplementary Fig. 6A, B. Scale bars: 2 µm. C Representative images of Co-IF against the Mbnl1 and Tdp-43 in myofibers cultured for 18 h in the presence of nocodazole or control (DMSO). Scale bars: 2 µm. D Perinuclear to cytoplasmic relative IF signal density in nocodazole (red) and control (DMSO, blue) myofibers. n = 5 myofibers per protein/condition. *p < 0.05, Two-sided Mann–Whitney U test.
Fig. 8. RNAs exhibit restricted diffusion and directed transport in myotubes.
A Schematic describing MS2 live-cell RNA imaging strategy. B Example image of a myotube expressing MS2-labeled RNA (top) and RNA trajectories (bottom) from a 53-s imaging time course. Scale bars: 2 µm. C Percentage of RNA trajectories in each motion category. D Diffusion coefficients for low-mobility (red) and high-mobility (green) diffusive tracks and example tracks from both groups. X-axis ticks for scale: 0.5 µm. E Distance and velocity for processive (blue) and crawling trajectories (purple) along with example tracks from both groups. X-axis ticks for scale: 0.5 µm. F Series of images showing RNA particle splitting and merging events. Colored triangles denote particle identities. Scale bars: 1 µm. G Example RNA tracks from a myotube expressing MS2-labeled RNA during a 50-min imaging time course. Scale bars: 2 µm. H Maximum distance traveled for each track from the 50-min (red) and 2-min (blue) imaging time courses. Tracks below dotted line (1 µm) are categorized as stationary. I Fraction of stationary tracks in 50-min and 2-min time courses.
Fig. 9. Computational simulation confirms that directed transport is required to disperse mRNA in myofibers.
A Diagram of RNA mobility states modeled in the simulation of RNA motion in myofibers. B Network diagram of the discrete-time Markov chain (DTMC) model developed to simulate RNA generation, motion, and decay. C Comparison of RNA localization patterns observed in 1000 h simulations of Polr2a RNA either with or without directed transport states. D Distance to nucleus measured for simulated Polr2a RNAs with (orange) or without (blue) directed transport states. Shown for comparison is the distribution from a simulation of Polr2a mRNAs in the high-mobility diffusion state (green) and a null distribution generated from randomly selected cytoplasmic coordinates (gray).
Similar articles
- MACF1 controls skeletal muscle function through the microtubule-dependent localization of extra-synaptic myonuclei and mitochondria biogenesis.
Ghasemizadeh A, Christin E, Guiraud A, Couturier N, Abitbol M, Risson V, Girard E, Jagla C, Soler C, Laddada L, Sanchez C, Jaque-Fernandez FI, Jacquemond V, Thomas JL, Lanfranchi M, Courchet J, Gondin J, Schaeffer L, Gache V. Ghasemizadeh A, et al. Elife. 2021 Aug 27;10:e70490. doi: 10.7554/eLife.70490. Elife. 2021. PMID: 34448452 Free PMC article. - [Myonuclear domain settings by microtubules and MACF1].
Ghasemizadeh A, Gache V. Ghasemizadeh A, et al. Med Sci (Paris). 2024 Nov;40 Hors série n° 1:64-68. doi: 10.1051/medsci/2024134. Epub 2024 Nov 18. Med Sci (Paris). 2024. PMID: 39555882 French. - Insights into Cell-Specific Functions of Microtubules in Skeletal Muscle Development and Homeostasis.
Lucas L, Cooper TA. Lucas L, et al. Int J Mol Sci. 2023 Feb 2;24(3):2903. doi: 10.3390/ijms24032903. Int J Mol Sci. 2023. PMID: 36769228 Free PMC article. Review. - Transient association of titin and myosin with microtubules in nascent myofibrils directed by the MURF2 RING-finger protein.
Pizon V, Iakovenko A, Van Der Ven PF, Kelly R, Fatu C, Fürst DO, Karsenti E, Gautel M. Pizon V, et al. J Cell Sci. 2002 Dec 1;115(Pt 23):4469-82. doi: 10.1242/jcs.00131. J Cell Sci. 2002. PMID: 12414993 - RNA trafficking in oligodendrocytes.
Carson JH, Cui H, Krueger W, Schlepchenko B, Brumwell C, Barbarese E. Carson JH, et al. Results Probl Cell Differ. 2001;34:69-81. doi: 10.1007/978-3-540-40025-7_5. Results Probl Cell Differ. 2001. PMID: 11288680 Review.
Cited by
- Skeletal Muscle Cell Growth Alters the Lipid Composition of Extracellular Vesicles.
Valentino TR, Rule BD, Mobley CB, Nikolova-Karakashian M, Vechetti IJ. Valentino TR, et al. Membranes (Basel). 2021 Aug 12;11(8):619. doi: 10.3390/membranes11080619. Membranes (Basel). 2021. PMID: 34436382 Free PMC article. - The molecular athlete: exercise physiology from mechanisms to medals.
Furrer R, Hawley JA, Handschin C. Furrer R, et al. Physiol Rev. 2023 Jul 1;103(3):1693-1787. doi: 10.1152/physrev.00017.2022. Epub 2023 Jan 5. Physiol Rev. 2023. PMID: 36603158 Free PMC article. Review. - Intracellular Membrane Contact Sites in Skeletal Muscle Cells.
Serano M, Perni S, Pierantozzi E, Laurino A, Sorrentino V, Rossi D. Serano M, et al. Membranes (Basel). 2025 Jan 14;15(1):29. doi: 10.3390/membranes15010029. Membranes (Basel). 2025. PMID: 39852269 Free PMC article. Review. - Scaling of nuclear numbers and their spatial arrangement in skeletal muscle cell size regulation.
Hansson KA, Eftestøl E. Hansson KA, et al. Mol Biol Cell. 2023 Jul 1;34(8):pe3. doi: 10.1091/mbc.E22-09-0424. Mol Biol Cell. 2023. PMID: 37339435 Free PMC article. Review. - Pan-cellular organelles and suborganelles-from common functions to cellular diversity?
Schieweck R, Götz M. Schieweck R, et al. Genes Dev. 2024 Mar 22;38(3-4):98-114. doi: 10.1101/gad.351337.123. Genes Dev. 2024. PMID: 38485267 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources