Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements - PubMed (original) (raw)
Bacterial chromosomal mobility via lateral transduction exceeds that of classical mobile genetic elements
Suzanne Humphrey et al. Nat Commun. 2021.
Abstract
It is commonly assumed that the horizontal transfer of most bacterial chromosomal genes is limited, in contrast to the frequent transfer observed for typical mobile genetic elements. However, this view has been recently challenged by the discovery of lateral transduction in Staphylococcus aureus, where temperate phages can drive the transfer of large chromosomal regions at extremely high frequencies. Here, we analyse previously published as well as new datasets to compare horizontal gene transfer rates mediated by different mechanisms in S. aureus and Salmonella enterica. We find that the horizontal transfer of core chromosomal genes via lateral transduction can be more efficient than the transfer of classical mobile genetic elements via conjugation or generalized transduction. These results raise questions about our definition of mobile genetic elements, and the potential roles played by lateral transduction in bacterial evolution.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
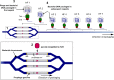
Fig. 1. Lateral transduction of chromosomal DNA.
LT is initiated during the early stages of prophage activation with the prophage remaining integrated in the bacterial chromosome. The mechanism commences with bidirectional in situ replication of the integrated prophage from the phage ori, generating multiple copies of the integrated phage and surrounding bacterial chromosome [1]. Some prophages subsequently excise from the chromosome to generate progeny via the lytic cycle, but in those that remain integrated, the phage small terminase subunit (TerS) recognises the embedded pac site (pink triangle) within the prophage sequence, forming a complex for delivery of the DNA to the large terminase (TerL) subunit [2]. The TerS:DNA complex associates with the large terminase (TerL) subunit, which cleaves and translocates the DNA into available phage capsids until capacity (one headful) is reached, with the initial capsid containing a mixture of phage and chromosomal DNA [3]. When the capsid is filled, the DNA is cleaved once more and the Terminase:DNA complex associates with a new empty capsid to resume the packaging process, generating many processive headfuls containing bacterial chromosomes for subsequent transduction [4].
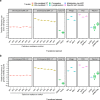
Fig. 2. Genetic mobility via different mechanisms in S. aureus.
a Transfer frequencies per donor cell and b relative frequency of genetic mobility (defined as transfer frequency × cargo capacity) of different genetic elements by transfer type. Transfer of chromosomal DNA by phage 80α via generalised (GT) or lateral (LT) transduction of a cadmium resistance marker located at defined distances from the phage attachment site, conjugative transposons (Conj. Tn), phages, PICIs and plasmids. Data were extracted from literature or acquired through experimentation (see Table 1 for details).
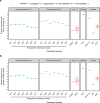
Fig. 3. Genetic mobility via different mechanisms in Salmonella spp.
a Transfer frequencies per donor cell and b relative frequency of genetic mobility (defined as transfer frequency × cargo capacity) of different genetic elements by transfer type. Transfer of chromosomal DNA by phage P22 via generalised (GT) or lateral (LT) transduction of a tetracycline resistance marker located at defined distances from the phage attachment site, conjugative transposons (Conj. Tn), phages and plasmids. Data were extracted from literature or acquired through experimentation (see Table 3 for details).
Fig. 4. Potential for extensive DNA transfer via lateral transduction is dictated by the distribution of prophage attachment sites in the bacterial chromosome.
Map of the chromosomes of S. aureus NCTC8325-4 (a) and S. Typhimurium LT2 (b) indicating the ten potential chromosomal attachment (attB) sites available for prophage integration. The packaging direction of an integrated prophage from each attB site is indicated by its corresponding colour-coded arrow, with the dashed regions representing a distance of approximately seven headfuls (one headful = ~45 kb for 80α) in S. aureus, or 12 headfuls (one headful = ~43 kb for P22) in S. Typhimurium from the attB site, which is the minimum distance known to be packaged by LT for each phage. a The distribution and directionality of attB sites in the S. aureus NCTC8325-4 chromosome indicates the potential for transfer of the entire region between ~0.31 and 2.0 Mb from a poly-lysogenic background, representing approximately 58% of the bacterial chromosome. Adapted from ref. . b The distribution and directionality of attB sites in S. Typhimurium LT2 indicates the potential for transfer of up to 3.5 Mb of chromosomal DNA from a poly-lysogenic background, representing approximately 72% of the bacterial chromosome. Source data are provided as a Source Data file.
Comment in
- Is the bacterial chromosome a mobile genetic element?
Hall JPJ. Hall JPJ. Nat Commun. 2021 Nov 4;12(1):6400. doi: 10.1038/s41467-021-26758-y. Nat Commun. 2021. PMID: 34737310 Free PMC article. - Laterally mobile chromosomes.
Clyde D. Clyde D. Nat Rev Genet. 2022 Jan;23(1):3. doi: 10.1038/s41576-021-00437-6. Nat Rev Genet. 2022. PMID: 34795384 No abstract available.
Similar articles
- Screening for Highly Transduced Genes in Staphylococcus aureus Revealed Both Lateral and Specialized Transduction.
Bowring JZ, Su Y, Alsaadi A, Svenningsen SL, Parkhill J, Ingmer H. Bowring JZ, et al. Microbiol Spectr. 2022 Feb 23;10(1):e0242321. doi: 10.1128/spectrum.02423-21. Epub 2022 Feb 9. Microbiol Spectr. 2022. PMID: 35138167 Free PMC article. - Bacteriophages of Staphylococcus aureus efficiently package various bacterial genes and mobile genetic elements including SCCmec with different frequencies.
Mašlaňová I, Doškař J, Varga M, Kuntová L, Mužík J, Malúšková D, Růžičková V, Pantůček R. Mašlaňová I, et al. Environ Microbiol Rep. 2013 Feb;5(1):66-73. doi: 10.1111/j.1758-2229.2012.00378.x. Epub 2012 Sep 3. Environ Microbiol Rep. 2013. PMID: 23757132 - Staphylococcus aureus mobile genetic elements.
Alibayov B, Baba-Moussa L, Sina H, Zdeňková K, Demnerová K. Alibayov B, et al. Mol Biol Rep. 2014 Aug;41(8):5005-18. doi: 10.1007/s11033-014-3367-3. Epub 2014 Apr 13. Mol Biol Rep. 2014. PMID: 24728610 Review. - Phage Transduction of Staphylococcus aureus.
Chu Yuan Kee MJ, Chen J. Chu Yuan Kee MJ, et al. Methods Mol Biol. 2024;2738:263-275. doi: 10.1007/978-1-0716-3549-0_17. Methods Mol Biol. 2024. PMID: 37966605 - Mechanisms of antibiotic resistance in Staphylococcus aureus.
Pantosti A, Sanchini A, Monaco M. Pantosti A, et al. Future Microbiol. 2007 Jun;2(3):323-34. doi: 10.2217/17460913.2.3.323. Future Microbiol. 2007. PMID: 17661706 Review.
Cited by
- Essential Topics for the Regulatory Consideration of Phages as Clinically Valuable Therapeutic Agents: A Perspective from Spain.
Vázquez R, Díez-Martínez R, Domingo-Calap P, García P, Gutiérrez D, Muniesa M, Ruiz-Ruigómez M, Sanjuán R, Tomás M, Tormo-Mas MÁ, García P. Vázquez R, et al. Microorganisms. 2022 Mar 26;10(4):717. doi: 10.3390/microorganisms10040717. Microorganisms. 2022. PMID: 35456768 Free PMC article. Review. - Chicken liver is a potential reservoir of bacteriophages and phage-derived particles containing antibiotic resistance genes.
Blanco-Picazo P, Gómez-Gómez C, Aguiló-Castillo S, Fernández-Orth D, Cerdà-Cuéllar M, Muniesa M, Rodríguez-Rubio L. Blanco-Picazo P, et al. Microb Biotechnol. 2022 Sep;15(9):2464-2475. doi: 10.1111/1751-7915.14056. Epub 2022 Apr 29. Microb Biotechnol. 2022. PMID: 35485188 Free PMC article. - Lateral transduction is inherent to the life cycle of the archetypical Salmonella phage P22.
Fillol-Salom A, Bacigalupe R, Humphrey S, Chiang YN, Chen J, Penadés JR. Fillol-Salom A, et al. Nat Commun. 2021 Nov 8;12(1):6510. doi: 10.1038/s41467-021-26520-4. Nat Commun. 2021. PMID: 34751192 Free PMC article. - Mechanisms of antimicrobial resistance in biofilms.
Liu HY, Prentice EL, Webber MA. Liu HY, et al. NPJ Antimicrob Resist. 2024;2(1):27. doi: 10.1038/s44259-024-00046-3. Epub 2024 Oct 1. NPJ Antimicrob Resist. 2024. PMID: 39364333 Free PMC article. Review. - Stable antibiotic resistance and rapid human adaptation in livestock-associated MRSA.
Matuszewska M, Murray GGR, Ba X, Wood R, Holmes MA, Weinert LA. Matuszewska M, et al. Elife. 2022 Jun 28;11:e74819. doi: 10.7554/eLife.74819. Elife. 2022. PMID: 35762208 Free PMC article.
References
- Siefert, J. L. Defining the Mobilome. in (eds Gogarten, M. B., Gogarten, J. P. & Olendzenski, L. C.) vol. 532, 13–27 (Humana Press, 2009). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- MR/M003876/1/MRC_/Medical Research Council/United Kingdom
- BB/V002376/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- 201531/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- MR/S00940X/2/MRC_/Medical Research Council/United Kingdom
- BB/N002873/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- WT_/Wellcome Trust/United Kingdom
- MR/S00940X/1/MRC_/Medical Research Council/United Kingdom
- MR/V000772/1 /MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources