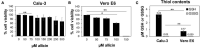The Effect of Allicin on the Proteome of SARS-CoV-2 Infected Calu-3 Cells - PubMed (original) (raw)
The Effect of Allicin on the Proteome of SARS-CoV-2 Infected Calu-3 Cells
Kirstin Mösbauer et al. Front Microbiol. 2021.
Abstract
Allicin (diallyl thiosulfinate) is the major thiol-reactive organosulfur compound produced by garlic plants (Allium sativum) upon tissue damage. Allicin exerts its strong antimicrobial activity against bacteria and fungi via _S_-thioallylation of protein thiols and low molecular weight thiols. Here, we investigated the effect of allicin on SARS-CoV-2 infected Vero E6 and Calu-3 cells. Toxicity tests revealed that Calu-3 cells showed greater allicin tolerance, probably due to >4-fold higher GSH levels compared to the very sensitive Vero E6 cells. Exposure of infected Vero E6 and Calu-3 cells to biocompatible allicin doses led to a ∼60-70% decrease of viral RNA and infectious viral particles. Label-free quantitative proteomics was used to investigate the changes in the Calu-3 proteome after SARS-CoV-2 infection and the effect of allicin on the host-virus proteome. SARS-CoV-2 infection of Calu-3 cells caused a strong induction of the antiviral interferon-stimulated gene (ISG) signature, including several antiviral effectors, such as cGAS, Mx1, IFIT, IFIH, IFI16, IFI44, OAS, and ISG15, pathways of vesicular transport, tight junctions (KIF5A/B/C, OSBPL2, CLTCL1, and ARHGAP17) and ubiquitin modification (UBE2L3/5), as well as reprogramming of host metabolism, transcription and translation. Allicin treatment of infected Calu-3 cells reduced the expression of IFN signaling pathways and ISG effectors and reverted several host pathways to levels of uninfected cells. Allicin further reduced the abundance of the structural viral proteins N, M, S and ORF3 in the host-virus proteome. In conclusion, our data demonstrate the antiviral and immunomodulatory activity of biocompatible doses of allicin in SARS-CoV-2-infected cell cultures. Future drug research should be directed to exploit the thiol-reactivity of allicin derivatives with increased stability and lower human cell toxicity as antiviral lead compounds.
Keywords: Calu-3; SARS-CoV-2; Vero E6; allicin; proteome.
Copyright © 2021 Mösbauer, Fritsch, Adrian, Bernhardt, Gruhlke, Slusarenko, Niemeyer and Antelmann.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
FIGURE 1
Human Calu-3 cells are more resistant to allicin compared to Vero E6. (A,B) Cell viability of untreated and allicin treated Calu-3 (A) and Vero E6 cells (B) was measured after 24 h using the CellTiter-Glo® Luminescent Cell Viability Assay (Promega) according to the manufacturer’s instructions. The viability of the control without allicin was set to 100%. The viability of Calu-3 cells was not significantly decreased upon exposure to 100–200 μM allicin, while concentrations of ≥100 μM allicin interfered with Vero E6 cell viability. (C) The levels of glutathione (GSH) and glutathione disulfide (GSSG) were determined in untreated Calu-3 and Vero E6 cells using the GSH/GSSG-Glo Assay (Promega) according to the manufacturer’s instructions. The results are from 4 biological replicates with 3 technical replicates for (C). Error bars represent the standard deviation (SD). _p_-values were calculated using an unpaired two-tailed _t_-test.*p < 0.05; **p < 0.01.
FIGURE 2
Allicin treatment of SARS-CoV-2 infected Vero E6 cells reduced the levels of infectious viral particles and viral RNA. (A–C) Vero E6 cells were infected with SARS-CoV-2 at a MOI of 0.01. After 24 h p.i. viral replication was analyzed by determination of infectious viral particles or viral RNA in the supernatant. (A) Comparison of different allicin treatments: Untreated Vero E6 cells infected with SARS-CoV-2 as control (SARS); SARS-CoV-2 pre-treated with 50 μM allicin for 30 min prior to infection of host cells (allicin + SARS); host cells pre-treated with 50 μM allicin for 30 min prior to infection with SARS-CoV-2 (allicin + Vero); and SARS-CoV-2 infected host cells treated with 50 μM allicin p.i. (allicin + Vero p.i.). (B,C) The amount of viral RNA (B) and infectious viral particles (C) was determined after treatment of SARS-CoV-2 infected Vero E6 cells with 50 and 75 μM allicin p.i. The results (A–C) are from three biological replicates with two technical replicates for panel (B). Error bars represent the SD. _p_-values were calculated using an unpaired two-tailed _t_-test.*p < 0.05; ***p < 0.001.
FIGURE 3
Allicin treatment of SARS-CoV-2 infected Calu-3 cells leads to decreased amounts of infectious viral particles and viral RNA. (A–D) SARS-CoV-2 infected Calu-3 cells were treated with 100 and 200 μM allicin p.i. The amount of viral RNA (A,B) and infectious viral particles (C,D) was determined 16 h (A,C) and 24 h (B,D) p.i. with SARS-CoV-2 at a MOI of 0.01 (A,C) and 0.005 (B,D). The results are from three biological replicates with two technical replicates for panels (A,B). Error bars represent the SD. _p_-values were calculated using an unpaired two-tailed _t_-test. *p < 0.05.
FIGURE 4
Cellular effect of 150 μM allicin on SARS-CoV-2 infected Calu-3 cells. Non-infected Calu-3 cells were cultivated for 24 h and served as controls (A). Calu-3 cells were infected with SARS-CoV-2 at a MOI of 0.01 and the SARS-CoV-2 induced cellular effects were studied at 24 h p.i. without (B) or with 150 μM allicin treatment (C) after infection as described in the “Materials and Methods” section. At 24 h p.i. infected Calu-3 cells showed cellular damages, including cell rounding, detachment and cell death (B). Allicin treatment decreased some cellular damages (C). Cells were imaged with a Nikon Ts2R-FL inverted microscope.
FIGURE 5
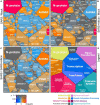
The effect of allicin on the proteome of SARS-CoV-2 infected Calu-3 cells. The host-viral proteome treemaps (A–C) show the 536 differentially expressed proteins upon SARS-CoV-2 infection and were constructed by the Paver software (Mehlan et al., 2013). The treemaps visualize the following proteome changes: (A) SARS-CoV-2 infection/Mock, (B) SARS-CoV-2 infection + Allicin/Mock and (C) SARS-CoV-2 infection −/+ Allicin. The treemap (D) serves as legend for the functional KEGG categories displayed in different colors for level 1 and sublevel 2 as listed in Supplementary Table 3. The cell sizes in panels (A–C) denote the average abundances of 207 proteins with ≥1.5-fold inductions and 329 proteins with <0.66-fold decreased expression after SARS-CoV-2 infection. The log2 ratios of the proteins are shown by a red-blue color gradient (red – induction, blue – repression) (A–C). _p_-values (p < 0.05; p < 0.01) were calculated using an unpaired two-tailed _t_-test from 3 to 4 biological replicates with 1–3 technical replicates (Supplementary Table 3).
FIGURE 6
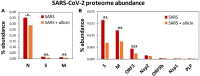
The effect of allicin on the abundance of SARS-CoV-2 proteins in the proteome of infected Calu-3 cells. (A,B) The abundance of the SARS-CoV-2 proteins (N, S, M, ORF3, Nsp2, ORF9b, Nsp1, and PLP) at 24 h p.i. relative to the total proteome abundance of infected Calu-3 cells was calculated in the absence (SARS) or presence of 150 μM allicin exposure p.i. (SARS + allicin). The N-protein showed a 21–29-fold higher abundance compared to the structural proteins S and M as shown in panel (A). The lower abundant S, M, ORF3, Nsp2, ORF9b, Nsp1, and PLP proteins are separately displayed in panel (B). The proteome results were obtained from 3–4 biological replicates with 1–3 technical replicates (Supplementary Table 3). Error bars represent the SD. _p_-values were calculated using an unpaired two-tailed _t_-test. *p < 0.05; **p < 0.01; ***p < 0.001.
FIGURE 7
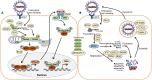
Schematic of viral RNA recognition, activation of the IFN and ISG signaling pathways (A) and antiviral functions of the identified ISG effectors (B) in this host-virus proteome study. (A) SARS-CoV-2 enters host cells via endocytosis. RIG-I is a cytosolic receptor to recognize viral RNA. cGAS and OAS are ISG effectors that function as RNA sensors. RIG-I, IFIH, and cGAS activate the mitochondrial antiviral-signaling protein (MAVS) and stimulator of IFN genes (STING), leading to phosphorylation of IFN responsive factors (e.g., IRF3), followed by IRF3 dimerization, translocation into the nucleus and transcriptional activation of IFN expression. IRF3 is negatively regulated by IFI44. RIG-I, MAVS, and STING can be regulated by ubiquitination (UBE2) and ISGylation (ISG15). Type-I IFN a/ß bind to the IFNAR receptor, resulting in phosphorylation of signal transducers and activators of transcription (STAT1/2) by the JAK and TYK kinases. Phosphorylated STAT1/2 form dimers and bind to IRF9, which triggers transcription of IFN-stimulated genes (ISGs) in the nucleus. (B) Antiviral ISG effectors affect different stages of the viral life cycle. Mx1 inhibits virus endocytosis and uncoating of the ribonucleocapsid. IFI16, OAS, and IFIT function in viral RNA degradation and block translation. IFIT, ISG15, TRIM, and UBE2 inhibit transcription, replication, translation or virus assembly. FKBP4 promotes protein folding. Kinesins (KIFA/B/C), Clathrin (CLTCL1), and TUBAL3 are involved in transport of virus vesicles. ARHGAP17 facilitates the formation or repair of tight junctions. The figure is adapted from reference (Schneider et al., 2014).
Similar articles
- Staphylococcus aureus responds to allicin by global S-thioallylation - Role of the Brx/BSH/YpdA pathway and the disulfide reductase MerA to overcome allicin stress.
Loi VV, Huyen NTT, Busche T, Tung QN, Gruhlke MCH, Kalinowski J, Bernhardt J, Slusarenko AJ, Antelmann H. Loi VV, et al. Free Radic Biol Med. 2019 Aug 1;139:55-69. doi: 10.1016/j.freeradbiomed.2019.05.018. Epub 2019 May 20. Free Radic Biol Med. 2019. PMID: 31121222 - The Disulfide Stress Response and Protein _S_-thioallylation Caused by Allicin and Diallyl Polysulfanes in Bacillus subtilis as Revealed by Transcriptomics and Proteomics.
Chi BK, Huyen NTT, Loi VV, Gruhlke MCH, Schaffer M, Mäder U, Maaß S, Becher D, Bernhardt J, Arbach M, Hamilton CJ, Slusarenko AJ, Antelmann H. Chi BK, et al. Antioxidants (Basel). 2019 Nov 29;8(12):605. doi: 10.3390/antiox8120605. Antioxidants (Basel). 2019. PMID: 31795512 Free PMC article. - The human allicin-proteome: S-thioallylation of proteins by the garlic defence substance allicin and its biological effects.
Gruhlke MCH, Antelmann H, Bernhardt J, Kloubert V, Rink L, Slusarenko AJ. Gruhlke MCH, et al. Free Radic Biol Med. 2019 Feb 1;131:144-153. doi: 10.1016/j.freeradbiomed.2018.11.022. Epub 2018 Nov 27. Free Radic Biol Med. 2019. PMID: 30500420 Free PMC article. - Allicin, the Odor of Freshly Crushed Garlic: A Review of Recent Progress in Understanding Allicin's Effects on Cells.
Borlinghaus J, Foerster Née Reiter J, Kappler U, Antelmann H, Noll U, Gruhlke MCH, Slusarenko AJ. Borlinghaus J, et al. Molecules. 2021 Mar 10;26(6):1505. doi: 10.3390/molecules26061505. Molecules. 2021. PMID: 33801955 Free PMC article. Review. - Paving New Roads Using Allium sativum as a Repurposed Drug and Analyzing its Antiviral Action Using Artificial Intelligence Technology.
Atoum MF, Padma KR, Don KR. Atoum MF, et al. Iran J Pharm Res. 2023 Jan 21;21(1):e131577. doi: 10.5812/ijpr-131577. eCollection 2022 Dec. Iran J Pharm Res. 2023. PMID: 36915406 Free PMC article. Review.
Cited by
- Assessment of Supplementation with Different Biomolecules in the Prevention and Treatment of COVID-19.
González-Acedo A, Manzano-Moreno FJ, García-Recio E, Ruiz C, Luna-Bertos E, Costela-Ruiz VJ. González-Acedo A, et al. Nutrients. 2024 Sep 11;16(18):3070. doi: 10.3390/nu16183070. Nutrients. 2024. PMID: 39339670 Free PMC article. Review. - Identification of new benzofuran derivatives as STING agonists with broad-spectrum antiviral activity.
Paulis A, Onali A, Vidalain PO, Lotteau V, Jaquemin C, Corona A, Distinto S, Delogu GL, Tramontano E. Paulis A, et al. Virus Res. 2024 Sep;347:199432. doi: 10.1016/j.virusres.2024.199432. Epub 2024 Jul 8. Virus Res. 2024. PMID: 38969014 Free PMC article. - Alterations in Circadian Rhythms, Sleep, and Physical Activity in COVID-19: Mechanisms, Interventions, and Lessons for the Future.
Das S, Khan R, Banerjee S, Ray S, Ray S. Das S, et al. Mol Neurobiol. 2024 May 3. doi: 10.1007/s12035-024-04178-5. Online ahead of print. Mol Neurobiol. 2024. PMID: 38702566 Review. - Systematic Review on Major Antiviral Phytocompounds from Common Medicinal Plants against SARS-CoV-2.
Ghosh S, Singha PS, Das LK, Ghosh D. Ghosh S, et al. Med Chem. 2024;20(6):613-629. doi: 10.2174/0115734064262843231120051452. Med Chem. 2024. PMID: 38317467 - Combating Black Fungus: Using Allicin as a Potent Antifungal Agent against Mucorales.
Schier C, Gruhlke MCH, Reucher G, Slusarenko AJ, Rink L. Schier C, et al. Int J Mol Sci. 2023 Dec 15;24(24):17519. doi: 10.3390/ijms242417519. Int J Mol Sci. 2023. PMID: 38139348 Free PMC article.
References
- Anderson T. W., Darling D. A. (1954). A test of goodness of fit. J. Am. Statist. Assoc. 49 765–769. 10.1080/01621459.1954.10501232 - DOI
LinkOut - more resources
Full Text Sources
Miscellaneous