Emergence of methicillin resistance predates the clinical use of antibiotics - PubMed (original) (raw)
. 2022 Feb;602(7895):135-141.
doi: 10.1038/s41586-021-04265-w. Epub 2022 Jan 5.
Claire L Raisen # 2, Xiaoliang Ba 2, Nicholas J Sadgrove 3, Guillermo F Padilla-González 3, Monique S J Simmonds 3, Igor Loncaric 4, Heidrun Kerschner 5, Petra Apfalter 5, Rainer Hartl 5, Ariane Deplano 6, Stien Vandendriessche 6 7, Barbora Černá Bolfíková 8, Pavel Hulva 9 10, Maiken C Arendrup 11, Rasmus K Hare 11, Céline Barnadas 11 12, Marc Stegger 11, Raphael N Sieber 11, Robert L Skov 13, Andreas Petersen 11, Øystein Angen 11, Sophie L Rasmussen 14 15, Carmen Espinosa-Gongora 16, Frank M Aarestrup 17, Laura J Lindholm 18, Suvi M Nykäsenoja 19, Frederic Laurent 20, Karsten Becker 21, Birgit Walther 22 23, Corinna Kehrenberg 24, Christiane Cuny 25, Franziska Layer 25, Guido Werner 25, Wolfgang Witte 25, Ivonne Stamm 26, Paolo Moroni 27 28, Hannah J Jørgensen 29, Hermínia de Lencastre 30 31, Emilia Cercenado 32, Fernando García-Garrote 32 33, Stefan Börjesson 34 35, Sara Hæggman 35, Vincent Perreten 36, Christopher J Teale 37, Andrew S Waller 38 39 40, Bruno Pichon 41, Martin D Curran 42, Matthew J Ellington 42 41, John J Welch 43, Sharon J Peacock 44, David J Seilly 2, Fiona J E Morgan 2 45, Julian Parkhill 2, Nazreen F Hadjirin 2, Jodi A Lindsay 46, Matthew T G Holden 47, Giles F Edwards 48, Geoffrey Foster 49, Gavin K Paterson 50, Xavier Didelot 51, Mark A Holmes 2, Ewan M Harrison 44 52 53, Anders R Larsen 11
Affiliations
- PMID: 34987223
- PMCID: PMC8810379
- DOI: 10.1038/s41586-021-04265-w
Emergence of methicillin resistance predates the clinical use of antibiotics
Jesper Larsen et al. Nature. 2022 Feb.
Abstract
The discovery of antibiotics more than 80 years ago has led to considerable improvements in human and animal health. Although antibiotic resistance in environmental bacteria is ancient, resistance in human pathogens is thought to be a modern phenomenon that is driven by the clinical use of antibiotics1. Here we show that particular lineages of methicillin-resistant Staphylococcus aureus-a notorious human pathogen-appeared in European hedgehogs in the pre-antibiotic era. Subsequently, these lineages spread within the local hedgehog populations and between hedgehogs and secondary hosts, including livestock and humans. We also demonstrate that the hedgehog dermatophyte Trichophyton erinacei produces two β-lactam antibiotics that provide a natural selective environment in which methicillin-resistant S. aureus isolates have an advantage over susceptible isolates. Together, these results suggest that methicillin resistance emerged in the pre-antibiotic era as a co-evolutionary adaptation of S. aureus to the colonization of dermatophyte-infected hedgehogs. The evolution of clinically relevant antibiotic-resistance genes in wild animals and the connectivity of natural, agricultural and human ecosystems demonstrate that the use of a One Health approach is critical for our understanding and management of antibiotic resistance, which is one of the biggest threats to global health, food security and development.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
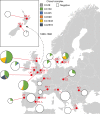
Fig. 1. Distribution of _mecC_-MRSA clones in European and New Zealand hedgehog samples.
The analysis included 828 samples from the nasal area, skin and feet of 276 hedgehogs originating from 16 wildlife rescue centres in 10 European countries and 2 wildlife rescue centres in New Zealand. The red dots indicate the sampling locations. The pie charts are connected to the sampling locations by a red line. The area of the pie chart is proportional to the number of samples from that location. The introduction of European hedgehogs into New Zealand from the UK between 1869 and 1892 is shown. A detailed description of the results is provided in Extended Data Fig. 1. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics. Source data
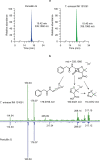
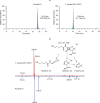
Fig. 2. Penicillin biosynthetic genes and antibiotic activity of T. erinacei IMI 101051.
a, Schematic of the key steps in the biosynthesis of penicillin G and cephalosporin C. The presence (green) or absence (red) of T. erinacei penicillin G and cephalosporin C biosynthetic genes is indicated. b, T . erinacei inhibition zones against a collection of S. aureus control strains (black) and two _mecC_-MRSA wild-type strains belonging to CC130 (green) and CC425 (blue) and their isogenic mutants. Two-tailed paired Student’s _t_-tests were used to compare inhibition zones of each mutant to the corresponding wild-type strain. Data are mean ± s.d.; n = 4 biologically independent fungal culture extracts. A detailed description of the results is provided in Extended Data Fig. 4. Source data
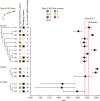
Fig. 3. Timeline of _mecC_-MRSA CC130, CC425 and CC1943 evolution in Europe.
Manual mapping of the tips on the type XI SCC_mec_ phylogeny onto the CC130, CC425 and CC1943 phylogenies, and vice versa, enabled us to assign the mecC_-MRSA isolates to 16 monophyletic lineages containing orthologous type XI SCC_mec elements (A–G). The trees are redrawn from Supplementary Figs. 2–5 to illustrate the branching order of the different type XI SCC_mec_ variants and _mecC_-MRSA lineages. Branch lengths are not drawn to scale. The presence and absence of hedgehog isolates in a given lineage are shown as black and white boxes, respectively. A detailed description of the geographical distribution and host range of major _mecC_-MRSA CC130, CC425 and CC1943 lineages is provided in Extended Data Fig. 7. The estimated date of the most recent common ancestor and 95% confidence interval of each _mecC_-MRSA lineage are illustrated by filled circles and horizontal lines, respectively. The introduction of penicillin G and methicillin as therapeutic options is indicated by red lines. Source data
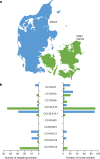
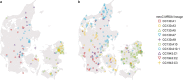
Fig. 4. Population structures of Danish _mecC_-MRSA isolates from hedgehogs and humans.
a, The map of Denmark shows the geographical ranges of two of the three hedgehog subpopulations in Jutland and on the major islands. b, The geographical distribution of major _mecC_-MRSA CC130, CC425 and CC1943 lineages in two broad collections of _mecC_-MRSA isolates recovered from hedgehogs (n = 141) and humans (n = 327) in Jutland and on the major islands. Hedgehog and human isolates from the remaining hedgehog subpopulation on the small island of Bornholm (not shown) were excluded from the analysis due to their small sample size (n = 9). A detailed map of the sampling locations is provided in Extended Data Fig. 8. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics. Source data
Extended Data Fig. 1. Distribution of _mecC_-MRSA clones in European and New Zealand hedgehogs.
The analysis included 828 samples from the nasal area, skin and feet of 276 hedgehogs originating from 16 wildlife rescue centres in ten European countries and two wildlife rescue centres in New Zealand. a, Presence of _mecC_-MRSA in hedgehogs (n = 276). Presence and absence are shown as black and white boxes, respectively. b, Distribution of _mecC_-MRSA clones in hedgehog samples (n = 828). c, Distribution of MSSA clones in MRSA-negative hedgehog samples (n = 606). Source data
Extended Data Fig. 2. Detection of penicillin G in T. erinacei IMI 101051 culture broth.
a, Left and right panels show extracted ion chromatogram of a pure standard of penicillin G and in T. erinacei culture broth, respectively. b, Upper and lower panels show MS2 spectra of penicillin G in a T. erinacei culture broth and a pure standard of penicillin G, respectively.
Extended Data Fig. 3. Detection of KPN in T. erinacei IMI 101051 culture broth.
a, Left and right panels show extracted ion chromatogram of a pure standard of penicillin G and KPN in T. erinacei culture broth, respectively. b, Upper and lower panels show MS2 spectra of KPN in a T. erinacei culture broth and a pure standard of penicillin G, respectively.
Extended Data Fig. 4. Antibiotic activity of T. erinacei IMI 101051.
T. erinacei inhibition zones against a collection of S. aureus control strains and two _mecC_-MRSA wild-type strains belonging to CC130 (02.5099.D) and CC425 (LGA251) and their isogenic mutants. The numbers on the plates refer to each of four biologically independent fungal culture extracts. Source data
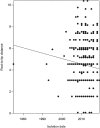
Extended Data Fig. 5. Root-to-tip linear regression analysis of the type XI SCC_mec_ dataset.
The correlation between root-to-tip distances and isolation dates is very weak with a coefficient of determination _R_2 = −0.05.
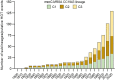
Extended Data Fig. 6. Number of _mecC_-MRSA CC1943 sublineages.
The bars show the number of sublineages of _mecC_-MRSA CC1943:C1, _mecC_-MRSA CC1943:C2 and _mecC_-MRSA CC1943:C3 at different time points. Source data
Extended Data Fig. 7. Geographical distribution and host range of major _mecC_-MRSA CC130, CC425 and CC1943 lineages.
The study collection included 991 _mecC_-MRSA CC130, CC425 and CC1943 isolates originating from 16 European countries. The countries of sampling and the eight largest (≥25 isolates) _mecC_-MRSA lineages are shown. Isolates belonging to the eight minor _mecC_-MRSA lineages are grouped together (others). The pie charts depict the proportion of _mecC_-MRSA isolates from hedgehogs (black), humans (grey) and other sources (white). The area of the pie chart is proportional to the number of _mecC_-MRSA isolates from that country. Source data
Extended Data Fig. 8. Sampling locations of Danish _mecC_-MRSA isolates from hedgehogs and humans.
The maps of Denmark relate to the map shown in Fig. 4. a, _mecC_-MRSA recovered from hedgehogs (n = 141). b, _mecC_-MRSA recovered from humans (n = 327). The location of each sample is given at the zip code area level. Maps were provided by Eurostat under a Creative Commons Attribution 4.0 International (CC BY 4.0) licence; the administrative boundaries are copyright of EuroGeographics.
Extended Data Fig. 9. Frequency of potential transmission events of _mecC_-MRSA CC130 isolates within wildlife rescue centres.
The bars show the proportion of isolates that belong to clusters defined at different pairwise SNP distance thresholds. Source data
Comment in
- Tracing the origins of antibiotic resistance.
Vandenbroucke-Grauls CMJE, Kluytmans JAJW. Vandenbroucke-Grauls CMJE, et al. Nat Med. 2022 Apr;28(4):638-640. doi: 10.1038/s41591-022-01752-z. Nat Med. 2022. PMID: 35318464 No abstract available.
Similar articles
- Benzylpenicillin-producing Trichophyton erinacei and methicillin resistant Staphylococcus aureus carrying the mecC gene on European hedgehogs - A pilot-study.
Dube F, Söderlund R, Lampinen Salomonsson M, Troell K, Börjesson S. Dube F, et al. BMC Microbiol. 2021 Jul 15;21(1):212. doi: 10.1186/s12866-021-02260-9. BMC Microbiol. 2021. PMID: 34266385 Free PMC article. - Stable antibiotic resistance and rapid human adaptation in livestock-associated MRSA.
Matuszewska M, Murray GGR, Ba X, Wood R, Holmes MA, Weinert LA. Matuszewska M, et al. Elife. 2022 Jun 28;11:e74819. doi: 10.7554/eLife.74819. Elife. 2022. PMID: 35762208 Free PMC article. - Methicillin-resistant Staphylococcus aureus emerged long before the introduction of methicillin into clinical practice.
Harkins CP, Pichon B, Doumith M, Parkhill J, Westh H, Tomasz A, de Lencastre H, Bentley SD, Kearns AM, Holden MTG. Harkins CP, et al. Genome Biol. 2017 Jul 20;18(1):130. doi: 10.1186/s13059-017-1252-9. Genome Biol. 2017. PMID: 28724393 Free PMC article. - Beta-lactam resistance in Staphylococcus aureus: the adaptive resistance of a plastic genome.
Fuda CC, Fisher JF, Mobashery S. Fuda CC, et al. Cell Mol Life Sci. 2005 Nov;62(22):2617-33. doi: 10.1007/s00018-005-5148-6. Cell Mol Life Sci. 2005. PMID: 16143832 Free PMC article. Review. - An Interplay of Multiple Positive and Negative Factors Governs Methicillin Resistance in Staphylococcus aureus.
Bilyk BL, Panchal VV, Tinajero-Trejo M, Hobbs JK, Foster SJ. Bilyk BL, et al. Microbiol Mol Biol Rev. 2022 Jun 15;86(2):e0015921. doi: 10.1128/mmbr.00159-21. Epub 2022 Apr 14. Microbiol Mol Biol Rev. 2022. PMID: 35420454 Free PMC article. Review.
Cited by
- Gut-Antimicrobial Peptides: Synergistic Co-Evolution with Antibiotics to Combat Multi-Antibiotic Resistance.
Baindara P, Mandal SM. Baindara P, et al. Antibiotics (Basel). 2023 Dec 14;12(12):1732. doi: 10.3390/antibiotics12121732. Antibiotics (Basel). 2023. PMID: 38136766 Free PMC article. Review. - Facing Danger: Exploring Personality and Reactions of European Hedgehogs (Erinaceus europaeus) towards Robotic Lawn Mowers.
Rasmussen SL, Schrøder BT, Berger A, Macdonald DW, Pertoldi C, Briefer EF, Alstrup AKO. Rasmussen SL, et al. Animals (Basel). 2023 Dec 19;14(1):2. doi: 10.3390/ani14010002. Animals (Basel). 2023. PMID: 38200733 Free PMC article. - First hip hemiarthroplasty in a Göttingen Minipig; surgical and post-mortem protocol.
Hartmann KT, Odgaard A, Knudsen UK, Aalbæk B, Kvich L, Birch JM, Petersen A, Bjarnsholt T, Jensen HE, Jensen LK. Hartmann KT, et al. J Orthop Surg Res. 2024 Sep 6;19(1):549. doi: 10.1186/s13018-024-05040-z. J Orthop Surg Res. 2024. PMID: 39243099 Free PMC article. - Molecular mechanisms of antibiotic resistance revisited.
Darby EM, Trampari E, Siasat P, Gaya MS, Alav I, Webber MA, Blair JMA. Darby EM, et al. Nat Rev Microbiol. 2023 May;21(5):280-295. doi: 10.1038/s41579-022-00820-y. Epub 2022 Nov 21. Nat Rev Microbiol. 2023. PMID: 36411397 Review. - A Review of the Occurrence of Metals and Xenobiotics in European Hedgehogs (Erinaceus europaeus).
Rasmussen SL, Pertoldi C, Roslev P, Vorkamp K, Nielsen JL. Rasmussen SL, et al. Animals (Basel). 2024 Jan 11;14(2):232. doi: 10.3390/ani14020232. Animals (Basel). 2024. PMID: 38254401 Free PMC article. Review.
References
- European Centre for Disease Prevention and Control, European Medicines Agencies. The Bacterial Challenge: Time to React. A Call to Narrow the Gap Between Multidrug-Resistant Bacteria in the EU and the Development of New Antibacterial Agentshttps://ecdc.europa.eu/sites/portal/files/media/en/publications/Publicat... (2009).
- Jevons MP. “Celbenin”—resistant Staphylococci. Br. Med. J. 1961;1:124–125.
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
- G1001787/MRC_/Medical Research Council/United Kingdom
- MR/N002660/1/MRC_/Medical Research Council/United Kingdom
- MR/P007201/1/MRC_/Medical Research Council/United Kingdom
- MR/S00291X/1/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical