Gut microbiome and health: mechanistic insights - PubMed (original) (raw)
Review
Gut microbiome and health: mechanistic insights
Willem M de Vos et al. Gut. 2022 May.
Abstract
The gut microbiota is now considered as one of the key elements contributing to the regulation of host health. Virtually all our body sites are colonised by microbes suggesting different types of crosstalk with our organs. Because of the development of molecular tools and techniques (ie, metagenomic, metabolomic, lipidomic, metatranscriptomic), the complex interactions occurring between the host and the different microorganisms are progressively being deciphered. Nowadays, gut microbiota deviations are linked with many diseases including obesity, type 2 diabetes, hepatic steatosis, intestinal bowel diseases (IBDs) and several types of cancer. Thus, suggesting that various pathways involved in immunity, energy, lipid and glucose metabolism are affected.In this review, specific attention is given to provide a critical evaluation of the current understanding in this field. Numerous molecular mechanisms explaining how gut bacteria might be causally linked with the protection or the onset of diseases are discussed. We examine well-established metabolites (ie, short-chain fatty acids, bile acids, trimethylamine N-oxide) and extend this to more recently identified molecular actors (ie, endocannabinoids, bioactive lipids, phenolic-derived compounds, advanced glycation end products and enterosynes) and their specific receptors such as peroxisome proliferator-activated receptor alpha (PPARα) and gamma (PPARγ), aryl hydrocarbon receptor (AhR), and G protein-coupled receptors (ie, GPR41, GPR43, GPR119, Takeda G protein-coupled receptor 5).Altogether, understanding the complexity and the molecular aspects linking gut microbes to health will help to set the basis for novel therapies that are already being developed.
Keywords: intestinal barrier function; intestinal microbiology; liver; obesity; probiotics.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY. Published by BMJ.
Conflict of interest statement
Competing interests: PDC and WMdV are inventors on patent applications dealing with the use of Akkermansia muciniphila and its components in the treatment of metabolic disorders. PDC and WMdV are co-founders of A-Mansia Biotech. PDC is co-founder of Enterosys. WMdV is co-founder of Caelus Health and inventor on patents on the use of Eubacterium hallii.
Figures
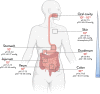
Figure 1
Total abundance of bacteria according to the different body sites. Bounds for bacteria number in different organs, derived from bacterial concentrations and volume.
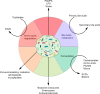
Figure 2
Molecules and metabolites produced by the gut microbiota according to the nutrients or metabolic source and their derived compounds. BSCFA, branched SCFA; LPS, lipopolysaccharides; PAMPs, pathogen-associated molecular patterns; SCFA, short chain fatty acids.
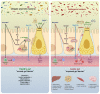
Figure 3
Molecular mechanisms linking gut microbiota and host health in both healthy and pathological situation. In healthy situation, colonocytes use butyrate as energy substrate via the beta-oxidation in the mitochondria, thereby consuming oxygen and directly contributing to maintain anaerobic condition in the lumen. Butyrate also binds to peroxisome proliferator-activated receptor gamma (PPARγ) which in turn repress inducible nitric oxide synthase (iNOS), decreases nitric oxide production (NO) and eventually nitrate production. Conversely, in pathological situations low butyrate content in the lumen is associated with lower PPARγ activity, increased glycolysis and lower oxygen consumption. This is associated with a higher expression of iNOS which in turn produces more NO and eventually increases nitrates availability for specific pathogens. Butyrate can also stimulate immune cells such as regulatory T cells (Treg) to reduce inflammation. The nuclear transcription factor aryl hydrocarbon receptor (AhR) is highly expressed and activated in healthy colonocytes, whereas agonists of AhR are lower or reduced AhR activity can lead to altered gut barrier function. Enteroendocrine cells (L-cells) are expressing several key receptors activated by short chain fatty acids (SCFAs), specific endocannabinoids (eCBs) and bile acids (BAs). Activating these receptors increase the secretion of key gut peptides such as glucagon-like peptide (GLP)-1, GLP-2 and peptide YY (PYY). Altogether, the interaction between the gut microbes and these molecular actors contributes to reduce intestinal permeability, to improve insulin secretion and insulin sensitivity, to reduce food intake, to lower plasma lipids and to avoid hepatic steatosis and metabolic endotoxaemia. All these effects are associated with lower inflammation. Conversely, opposite effects have been observed in pathological situations.
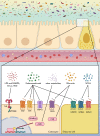
Figure 4
Colonocytes and endocrine cells express a variety of receptors able to sense and transmit signals from the microbial environment. Microbial/Pathogen-associated molecular patterns (PAMPs), lipopolyscaccharides (LPS) from the microbiota are detected by pattern recognition receptors, including toll-like receptors (TLRs). Amuc_1100 is a protein expressed on the outer membrane of Akkermansia muciniphila and which has been shown to signal through TLR2 to improve gut barrier function and reduce inflammation. Metabolites secreted by certain microbes (eg, endocannabinoids (eCBs)), generated by microbial digestion of dietary components (eg, short chain fatty acids (SCFAs)) or by transformation of host-derived factors (eg, eCBs and bile acids) can be sensed through various receptors and pathways to alter intestinal integrity and host health. CB1, CB2, cannabinoid receptor type 1 and type 2; TRPV1, transient receptor potential cation channel subfamily V member 1; FXR, farnesoid X receptor; AhR, aryl hydrocarbon receptor; GPR119, GPR43, GPR41, G-protein coupled receptor 119, 43 and 41; MYD88, myeloid differentiation primary response 88; PPARα/γ, peroxisome proliferator-activated receptors alpha and gamma; TGR5, Takeda G protein-coupled receptor 5.
Similar articles
- Impact of Bacterial Metabolites on Gut Barrier Function and Host Immunity: A Focus on Bacterial Metabolism and Its Relevance for Intestinal Inflammation.
Gasaly N, de Vos P, Hermoso MA. Gasaly N, et al. Front Immunol. 2021 May 26;12:658354. doi: 10.3389/fimmu.2021.658354. eCollection 2021. Front Immunol. 2021. PMID: 34122415 Free PMC article. Review. - The contribution of gut bacterial metabolites in the human immune signaling pathway of non-communicable diseases.
Hosseinkhani F, Heinken A, Thiele I, Lindenburg PW, Harms AC, Hankemeier T. Hosseinkhani F, et al. Gut Microbes. 2021 Jan-Dec;13(1):1-22. doi: 10.1080/19490976.2021.1882927. Gut Microbes. 2021. PMID: 33590776 Free PMC article. Review. - Interaction of gut microbiota with dysregulation of bile acids in the pathogenesis of nonalcoholic fatty liver disease and potential therapeutic implications of probiotics.
Chen J, Thomsen M, Vitetta L. Chen J, et al. J Cell Biochem. 2019 Mar;120(3):2713-2720. doi: 10.1002/jcb.27635. Epub 2018 Nov 15. J Cell Biochem. 2019. PMID: 30443932 Review. - Understanding connections and roles of gut microbiome in cardiovascular diseases.
Rajendiran E, Ramadass B, Ramprasath V. Rajendiran E, et al. Can J Microbiol. 2021 Feb;67(2):101-111. doi: 10.1139/cjm-2020-0043. Epub 2020 Oct 20. Can J Microbiol. 2021. PMID: 33079568 Review. - Metabolites Linking the Gut Microbiome with Risk for Type 2 Diabetes.
Zhu T, Goodarzi MO. Zhu T, et al. Curr Nutr Rep. 2020 Jun;9(2):83-93. doi: 10.1007/s13668-020-00307-3. Curr Nutr Rep. 2020. PMID: 32157661 Free PMC article. Review.
Cited by
- Multifaceted paternal exposures before conception and their epigenetic impact on offspring.
Wu X, Zhang W, Chen H, Weng J. Wu X, et al. J Assist Reprod Genet. 2024 Nov;41(11):2931-2951. doi: 10.1007/s10815-024-03243-1. Epub 2024 Sep 4. J Assist Reprod Genet. 2024. PMID: 39230664 Review. - A systematic framework for understanding the microbiome in human health and disease: from basic principles to clinical translation.
Ma Z, Zuo T, Frey N, Rangrez AY. Ma Z, et al. Signal Transduct Target Ther. 2024 Sep 23;9(1):237. doi: 10.1038/s41392-024-01946-6. Signal Transduct Target Ther. 2024. PMID: 39307902 Free PMC article. Review. - Regulatory effects of tea polysaccharides on hepatic inflammation, gut microbiota dysbiosis, and serum metabolomic signatures in beef cattle under heat stress.
Li F, Xu J, Xie M, Fei D, Zhou Y, Li X, Guang Y, Gong L, Hu L, Feng F. Li F, et al. Front Physiol. 2024 Sep 6;15:1460414. doi: 10.3389/fphys.2024.1460414. eCollection 2024. Front Physiol. 2024. PMID: 39308975 Free PMC article. - Difference in the Intestinal Microbiota between Breastfeed Infants and Infants Fed with Artificial Milk: A Systematic Review.
Inchingolo F, Inchingolo AM, Latini G, Ferrante L, de Ruvo E, Campanelli M, Longo M, Palermo A, Inchingolo AD, Dipalma G. Inchingolo F, et al. Pathogens. 2024 Jun 24;13(7):533. doi: 10.3390/pathogens13070533. Pathogens. 2024. PMID: 39057760 Free PMC article. Review. - Role of gut microbiota and immune cells in metabolic-associated fatty liver disease: clinical impact.
Alisi A, McCaughan G, Grønbæk H. Alisi A, et al. Hepatol Int. 2024 Oct;18(Suppl 2):861-872. doi: 10.1007/s12072-024-10674-6. Epub 2024 Jul 12. Hepatol Int. 2024. PMID: 38995341 Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical