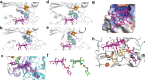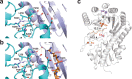Crystallographic snapshots of a B12-dependent radical SAM methyltransferase - PubMed (original) (raw)
Crystallographic snapshots of a B12-dependent radical SAM methyltransferase
Cameron D Fyfe et al. Nature. 2022 Feb.
Abstract
By catalysing the microbial formation of methane, methyl-coenzyme M reductase has a central role in the global levels of this greenhouse gas1,2. The activity of methyl-coenzyme M reductase is profoundly affected by several unique post-translational modifications3-6, such as a unique C-methylation reaction catalysed by methanogenesis marker protein 10 (Mmp10), a radical S-adenosyl-L-methionine (SAM) enzyme7,8. Here we report the spectroscopic investigation and atomic resolution structure of Mmp10 from Methanosarcina acetivorans, a unique B12 (cobalamin)-dependent radical SAM enzyme9. The structure of Mmp10 reveals a unique enzyme architecture with four metallic centres and critical structural features involved in the control of catalysis. In addition, the structure of the enzyme-substrate complex offers a glimpse into a B12-dependent radical SAM enzyme in a precatalytic state. By combining electron paramagnetic resonance spectroscopy, structural biology and biochemistry, our study illuminates the mechanism by which the emerging superfamily of B12-dependent radical SAM enzymes catalyse chemically challenging alkylation reactions and identifies distinctive active site rearrangements to provide a structural rationale for the dual use of the SAM cofactor for radical and nucleophilic chemistry.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
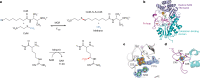
Fig. 1. MCR and Mmp10 activity with overall structure of Mmp10.
a, The activity of MCR producing CoB–CoM heterodisulfide and methane is enhanced by the post-translational modification of R285 catalysed by the B12-dependent radical SAM enzyme Mmp10. b, Overall structure of Mmp10 with bound sodium, [4Fe–4S] cluster, MTA, MeCbl and single iron atom cofactors (Protein Data Bank (PDB) accession 7QBT). Although Mmp10 was crystallized with SAM, only electron density for MTA was observed (Extended Data Fig. 1, Extended Data Table 1). c, Magnified view of the [4Fe–4S] cluster coordinated by three cysteine residues and Y115 alongside the modelled MTA molecule not coordinated to the cluster (Extended Data Fig. 1b). d, Iron loop with a single iron atom coordinated by four cysteine residues (Extended Data Fig. 1c). Light blue, radical SAM domain; teal, cobalamin-binding domain; purple, iron loop; green, MTA; magenta, MeCbl. The [4Fe–4S] cluster is shown as yellow and orange spheres; the single iron is presented as an orange sphere; and the sodium atom is shown as a violet sphere. Omit maps (blue mesh) of the [4Fe–4S] cluster, its coordinated Y115 and the uncoordinated MTA (c) or single iron atom (d) contoured at 3_σ_ are depicted.
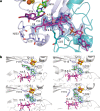
Fig. 2. Binding of vitamin B12 and _S_-adenosyl ligands by Mmp10.
a, The C8 side chain of MeCbl is shown in interaction with Y23, I220, L221 and N223 within the radical SAM domain, resulting in a planar tetrapyrrole ring. MeCbl has no lower axial ligand because it is pentacoordinated; however, L322, which resides at 3.9 Å from the cobalt atom, is part of a loop of residues forming a hydrophobic environment for the cobalt ion. Y23 appears at 4.8 Å from the cobalt ion. b,Snapshots of S_-adenosyl cofactors within distinct Mmp10 structures. The distances between the sulfur atom of SAH and MTA or SAM and the cobalt ion are indicated by dashed lines. Top left, Mmp10 crystallized with SAH in the absence of peptide substrate (1: Mmp10 SAH structure; PDB 7QBV). Bottom left and top right, Mmp10 crystallized in the absence of substrate with SAM. Only the density of MTA was observed, which is labelled accordingly (2: Mmp10 MTA_1; PDB 7QBT; 3: Mmp10 MTA_2; PDB 7QBU). Bottom right, Mmp10 crystallized with SAM and its peptide substrate (4: Mmp10–SAM–peptide structure; PDB 7QBS). Light blue and purple, radical SAM domain residues; teal, cobalamin domain; green, SAM, MTA and SAH; magenta, MeCbl. The [4Fe–4S] cluster is shown as orange and yellow spheres. Omit maps (blue mesh) of ligands contoured at 3_σ are depicted. See Extended Data Table 1 and Extended Data Fig. 4 for additional information.
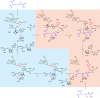
Fig. 3. Structure of Mmp10 in complex with its peptide substrate.
a, Overview of Mmp10 in complex with peptide substrate, shown in orange (PDB code 7QBS). b, Close-up of the Mmp10 active site showing SAM in green, peptide substrate in orange and cobalamin in magenta. The distance between the C5′ atom of SAM and the Cδ atom of the arginine peptide substrate R285 (3.7 Å) and that between R285 and the methyl group of MeCbl (4.1 Å) are indicated by dashed lines. The omit map (blue mesh) of peptide is contoured at 3_σ_. c, Sequences of the peptides used as potential substrates with the substitution of arginine (R285) with isoleucine (Ile), leucine (Leu), ornithine (Orn), lysine (Lys), citrulline (Cit) or homoarginine (HArg). d, UV–visible light analysis of Mmp10 pre-incubated with OHCbl. Green line, OHCbl–Mmp10; blue line, OHCbl–Mmp10 after incubation with Ti(III) citrate; red line, reduced OHCbl–Mmp10 exposed to air. See Extended Data Figs. 4–6 for additional information.
Fig. 4. Proposed mechanism for Mmp10.
Nucleophilic and radical catalysis are highlighted in blue and orange, respectively.
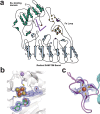
Extended Data Fig. 1. Topology diagram of Mmp10 depicting domains (a) and 2Fo-Fc and Fo-Fc maps of (b) SAM modeled as MTA, [4Fe-4S] cluster, Tyrosine 115 and (c) the single Fe ion in Mmp10 structure (Mmp10 MTA_1 structure).
Three yellow circles surrounding the [4Fe-4S] cluster represent cysteine residues that coordinate the radical SAM cluster. Purple circle represents Tyrosine 115 that coordinates the fourth iron of the [4Fe-4S] cluster. Four yellow circles within the Fe-loop represent cysteine residues that coordinate a single iron atom shown as an orange circle. Red dot represents Leucine 322 that provides a hydrophobic pocket at the lower axial position of the cobalamin cofactor. Purple sticks represent cobalamin. Pink dot depicts Glutamate 378 involved in substrate binding. Blue dot shows the sodium ion. The 2mFo-DFc map is colored in blue and contoured at 1.0 σ. The mFo-DFc map is colored green (3.0 σ) and red (−3.0 σ).
Extended Data Fig. 2. Sequence alignment of Mmp10 homologs.
258 sequences from the megacluster 1-7 (radicalsam.org), were aligned, and strictly conserved residues are highlighted in colors. For clarity, only eight representative sequences with various degree of similarity from 100 to 37% (indicated in the right column) are shown. The three cysteine residues coordinating the radical SAM cluster, the four cysteine residues coordinating the Fe-loop and Y115 are strictly conserved. Similarly, the residues involved in the B12-binding motif are highly conserved, notably the key L322.
Extended Data Fig. 3. EPR analysis and activity of Mmp10 and mutants.
a, LC-MS analysis of the reaction catalyzed by Mmp10. SAM [M]+= 399.14, 5′-dA [M+H]+= 252.11 and SAH [M+H]+= 385.13 are indicated. The peptide substrate [M+H]+= 1496.77 is converted into a methylated species [M+H] += 1510.79 (see panel c for full assignment). b, Time course analysis of the reaction catalyzed by Mmp10. c, LC-MS/MS analysis of the 13-mer peptide before (upper panel) and after (lower panel) incubation with Mmp10. See Extended Data Table 2 for full assignment. d, Schematic representation of the A3-mutant, biochemical characterization and EPR analysis. Right upper panel: LC-MS analysis of the reaction before (blue trace) and after (red trace) 1 hr incubation. Right lower panel: Time course analysis of the reaction. Left lower panel: CW EPR analysis. The weak signal measured at g ~ 2.0 represents adventitious FeS cluster. e, Schematic representation, EPR analysis and activity assay of the A4-mutant lacking C35, C38, C45 and C48 involved in the Fe-loop coordination. Upper right panel: CW EPR analysis. Lower panel: LC-MS analysis of the reaction catalyzed by the A4-mutant before (blue trace) and after (red trace) 1 hr incubation. SAM [M]+= 399.14, 5′-dA [M+H]+= 252.11 and SAH [M+H]+= 385.13 are indicated. f, Schematic representation of the C38A-mutant and biochemical characterization. Middle panels: LC-MS analysis of the reaction before (blue trace) and after (red trace) 60 min incubation. SAM [M]+= 399.14, 5′-dA [M+H]+= 252.11 and SAH [M+H]+= 385.13 are indicated. Right panel: Time course analysis of the reaction. g, Schematic representation of the Y115A-mutant and biochemical characterization. Right and middle panels: LC-MS analysis of the reaction before (blue trace) and after (red trace) 1 hr incubation. SAM [M]+= 399.14, 5′-dA [M+H]+= 252.11 and SAH [M+H]+= 385.13 are indicated. h, Schematic representation of the Y115F-mutant and biochemical characterization. Activity of the Y115F mutant in the absence (Left panel) or the presence (middle panel) of the peptide substrate. Right panel: LC-MS analysis of the reaction after 1 hr. All reactions were performed under anaerobic and reducing conditions with reconstituted enzymes and analyzed by LC-MS. Methylated peptide [M+H]+= 1510.79 (●,◯), 5′-dA [M+H]+= 252.11 (■, ☐) and SAH [M+H]+= 385.13 (▲, Δ) are indicated. Experiments were performed in duplicate. i, Schematic representation and EPR analysis of Mmp10. Ox: Oxidized enzyme as purified, Red: Reduced enzyme after anaerobic reconstitution and incubation with sodium dithionite. SAM: Reduced enzyme after incubation with SAM. g-values are indicated on the panels. In all panels, the radical SAM cluster ligated by Tyr-115, Cys-15, Cys-19 & Cys-22, the iron-loop coordinated by Cys-35, Cys-38, Cys-45 and Cys-48 and the cobalamin cofactor are indicated.
Extended Data Fig. 4. Methylcobalamin, MTA, SAH and SAM in Mmp10 structures.
2Fo-Fc and Fo-Fc maps of Mmp10 crystallized with SAH in the absence of peptide substrate (a: Mmp10 SAH structure). Mmp10 crystallized in the absence of substrate with SAM. Only the density of MTA was observed and labeled accordingly (b & c: Mmp10 MTA_1 and d: Mmp10 MTA_2 structures). Mmp10 crystallized with SAM and its peptide substrate (e: Mmp10 SAM peptide structure). The 2mFo-DFc map is colored blue and contoured at 1.0 σ. The mFo-DFc map is colored green (3.0 σ) and red (−3.0 σ). See Extended Data Table 1 & Fig. 2 for additional information. f, Cobalamin bound within Mmp10 (left; PDB 7QBT) and TsrM (right: PDB 6WTF). Cobalamin from TsrM shown in green and methylcobalamin from Mmp10 shown in magenta. The corrin ring and the sidechain C8 exhibit distinct conformations. g & h, Hydrophobic pocket of Mmp10 binding methylcobalamin (Mmp10 MTA_1 structure). The pocket directly beneath the cobalt of methylcobalamin has no water molecules and no charged sidechains, resulting in a hydrophobic environment. The distance between L322 and the cobalt ion is of 4 Å. Surface charge generated using APBS surface charge within PyMOL (Schrödinger).
Extended Data Fig. 5. Mmp10 interaction with its substrate.
a, Structure superimposition of substrate-free with SAH and peptide-bound with SAM structures of Mmp10. b, Enlargement showing the major structural movements including the α1a helix and the α1 to α4 helices of the TIM barrel. Mmp10 with peptide bound is shown in grey and without peptide in teal. Structures were aligned according to the B12 binding domain. Alignment of the peptide-bound and substrate-free SAH structures using all domains (403 residues) has a r.m.s.d. of 0.963 Å. c, Polar interactions between Mmp10, its cofactors (methylcobalamin & SAM) and peptide substrate. Omit map of peptide substrate contoured at 3 σ level (in blue). SAM is shown in green, methylcobalamin is depicted in magenta, bound peptide is shown in orange, polar contacts are shown as grey dotted lines, water is shown as a red sphere. d, Omit map of the peptide substrate in Mmp10 active site contoured at 3 σ level (in light blue). e, The 2mFo-DFc map of the peptide substrate in Mmp10 active site is colored blue and contoured at 1.0 σ. The mFo-DFc map is colored green (3.0 σ) and red (−3.0 σ). SAM shown in green, methylcobalamin depicted in magenta, peptide substrate in orange.
Extended Data Fig. 6. Activity of Mmp10 on peptide substrate.
a, Influence of the critical R285 residue. Reactions were analyzed by LC-MS. Only for the wild-type peptide a methyl transfer reaction was observed (mass shift of Δm= +14 Da). See Extended Data Table 2 for peptide masses and Extended methods for reaction conditions. b, Activity of Mmp10 incubated with the wild-type peptide alone (left panel) or the wild-type and citrulline-containing peptides (right panel). Reactions were analyzed by LC-MS. Methylated peptide [M+H] += 1510.79 (circles), 5′-dA [M+H] += 252.11 (squares) and SAH [M+H] += 385.13 (triangles). Experiments were performed in duplicate. c, Formation of MeCbl by Mmp10 in the absence or the presence of Ti(III)citrate. Mmp10 was incubated under anaerobic and reducing conditions with SAM (1 mM), OHCbl (100 µM) and DTT (6 mM). Diamonds: Reaction with 1mM Ti(III)citrate. Squares: Reaction without Ti(III)citrate. Circles: Control reaction without enzyme and Ti(III)citrate. Experiments were performed in duplicate. MeCbl was detected by LC-MS analysis [M+H]2+: 672.80 and comparison with its retention time with authentic standard.
Extended Data Fig. 7. Fe loop and cation site in Mmp10.
a, Fe loop in Mmp10 (Mmp10 MTA_1 structure). Cobalamin colored magenta, residues from Fe loop colored purple and residue from cobalamin binding domain colored teal. b, Cation site in Mmp10 (Mmp10 MTA_1 structure). Na ion (purple sphere) modelled in cation site with an average distance of 2.4 Å in octahedral conformation (grey dotted lines) from coordinating sidechains and water. Distance between Na ion and 4Fe4S cluster is 11 Å (yellow dotted line). Omit map of Na ion contoured at 3 σ level. 4Fe4S cluster shown as orange and yellow spheres next to SAM molecule depicted in green. c, Activity of the D156A mutant (upper panel) compared to the WT enzyme (lower panel). Reactions were analyzed by LC-MS. Methylated peptide [M+H] += 1510.79 (circles), 5′-dA [M+H] += 252.11 (squares) and SAH [M+H] += 385.13 (triangles). Experiments were performed in duplicate.
Extended Data Fig. 8. Cis-peptide bond and metallic centers in Mmp10.
a, Cis-peptide bonds at the interface between the radical SAM and the cobalamin binding domains in the substrate free (Mmp10 MTA_1 structure) and b, peptide-bound Mmp10 structures. Peptide substrate shown in orange with the radical SAM domain coloured in light blue and cobalamin binding domain in teal. Polar interactions shown as yellow staggered lines. Cis-peptide bond between T257 and P258 are coordinating a water molecule which is also involved in substrate binding. The rare non-proline cis-peptide bond found between L259 and E260 is held in place through interactions between the sidechain nitrogens of R243 and R298 and the side chain oxygens of E260. c, Metallic centres in Mmp10. Mmp10 SAM peptide structure. Peptide substrate shown in orange.
Similar articles
- Methanogenesis marker protein 10 (Mmp10) from Methanosarcina acetivorans is a radical _S_-adenosylmethionine methylase that unexpectedly requires cobalamin.
Radle MI, Miller DV, Laremore TN, Booker SJ. Radle MI, et al. J Biol Chem. 2019 Aug 2;294(31):11712-11725. doi: 10.1074/jbc.RA119.007609. Epub 2019 May 20. J Biol Chem. 2019. PMID: 31113866 Free PMC article. - A Cobalamin-Dependent Radical SAM Enzyme Catalyzes the Unique Cα -Methylation of Glutamine in Methyl-Coenzyme M Reductase.
Gagsteiger J, Jahn S, Heidinger L, Gericke L, Andexer JN, Friedrich T, Loenarz C, Layer G. Gagsteiger J, et al. Angew Chem Int Ed Engl. 2022 Aug 8;61(32):e202204198. doi: 10.1002/anie.202204198. Epub 2022 Jun 29. Angew Chem Int Ed Engl. 2022. PMID: 35638156 Free PMC article. - Surprise! A hidden B12 cofactor catalyzes a radical methylation.
Jarrett JT. Jarrett JT. J Biol Chem. 2019 Aug 2;294(31):11726-11727. doi: 10.1074/jbc.H119.009976. J Biol Chem. 2019. PMID: 31375551 Free PMC article. - Radical-mediated enzymatic methylation: a tale of two SAMS.
Zhang Q, van der Donk WA, Liu W. Zhang Q, et al. Acc Chem Res. 2012 Apr 17;45(4):555-64. doi: 10.1021/ar200202c. Epub 2011 Nov 18. Acc Chem Res. 2012. PMID: 22097883 Free PMC article. Review. - Radical SAM enzymes in methylation and methylthiolation.
Hutcheson RU, Broderick JB. Hutcheson RU, et al. Metallomics. 2012 Nov;4(11):1149-54. doi: 10.1039/c2mt20136d. Epub 2012 Sep 19. Metallomics. 2012. PMID: 22992596 Free PMC article. Review.
Cited by
- Structural, Biochemical, and Bioinformatic Basis for Identifying Radical SAM Cyclopropyl Synthases.
Lien Y, Lachowicz JC, Mendauletova A, Zizola C, Ngendahimana T, Kostenko A, Eaton SS, Latham JA, Grove TL. Lien Y, et al. ACS Chem Biol. 2024 Feb 16;19(2):370-379. doi: 10.1021/acschembio.3c00583. Epub 2024 Jan 31. ACS Chem Biol. 2024. PMID: 38295270 - Structural and mechanistic basis for RiPP epimerization by a radical SAM enzyme.
Kubiak X, Polsinelli I, Chavas LMG, Fyfe CD, Guillot A, Fradale L, Brewee C, Grimaldi S, Gerbaud G, Thureau A, Legrand P, Berteau O, Benjdia A. Kubiak X, et al. Nat Chem Biol. 2024 Mar;20(3):382-391. doi: 10.1038/s41589-023-01493-1. Epub 2023 Dec 29. Nat Chem Biol. 2024. PMID: 38158457 - Changing Fates of the Substrate Radicals Generated in the Active Sites of the B12-Dependent Radical SAM Enzymes OxsB and AlsB.
Lee YH, Yeh YC, Fan PH, Zhong A, Ruszczycky MW, Liu HW. Lee YH, et al. J Am Chem Soc. 2023 Feb 15;145(6):3656-3664. doi: 10.1021/jacs.2c12953. Epub 2023 Jan 31. J Am Chem Soc. 2023. PMID: 36719327 Free PMC article. - Metabolic turnover of cysteine-related thiol compounds at environmentally relevant concentrations by Geobacter sulfurreducens.
Gutensohn M, Schaefer JK, Maas TJ, Skyllberg U, Björn E. Gutensohn M, et al. Front Microbiol. 2023 Jan 11;13:1085214. doi: 10.3389/fmicb.2022.1085214. eCollection 2022. Front Microbiol. 2023. PMID: 36713222 Free PMC article. - Structure and Catalytic Mechanism of Radical SAM Methylases.
Nguyen TQ, Nicolet Y. Nguyen TQ, et al. Life (Basel). 2022 Oct 28;12(11):1732. doi: 10.3390/life12111732. Life (Basel). 2022. PMID: 36362886 Free PMC article. Review.
References
- Conrad R. The global methane cycle: recent advances in understanding the microbial processes involved. Env. Microbiol. Rep. 2009;1:285–292. - PubMed
- Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Crystal structure of methyl-coenzyme M reductase: the key enzyme of biological methane formation. Science. 1997;278:1457–1462. - PubMed
- Kahnt J, et al. Post-translational modifications in the active site region of methyl-coenzyme M reductase from methanogenic and methanotrophic archaea. FEBS J. 2007;274:4913–4921. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous