The lncRNAs/miR-30e/CHI3L1 Axis Is Dysregulated in Systemic Sclerosis - PubMed (original) (raw)
The lncRNAs/miR-30e/CHI3L1 Axis Is Dysregulated in Systemic Sclerosis
Valentin Dichev et al. Biomedicines. 2022.
Abstract
Systemic sclerosis (SSc) is an autoimmune disease with completely undefined etiology and treatment difficulties. The expression of both protein coding and non-coding RNAs is dysregulated during disease development. We aimed to examine a possible regulatory axis implemented in the control of chitinase-3 like protein 1 (CHI3L1) or YKL-40, an inflammation-associated glycoprotein, shown to be elevated in SSc. A panel of seven miRNAs and three lncRNAs potentially involved in the control of CHI3L1 were selected on the basis of in silico analysis. TagMan assay was used to evaluate the expression levels of miRNAs and RT-qPCR for lncRNAs in white blood cells (WBCs) and plasma from SSc patients and healthy controls. Among the eight screened miRNAs, miR-30e-5p (p = 0.04) and miR-30a-5p (p = 0.01) were significantly downregulated in WBCs and plasma of SSc patients, respectively. On the contrary, the expression of the metastasis associated lung adenocarcinoma transcript 1 (MALAT1) (p = 0.044) and the Nuclear enriched abundant transcript 1 (NEAT1) (p = 0.008) in WBCs was upregulated compared to the controls. Increased levels of MALAT1 and NEAT1 could be associated with the downregulation of miR-30e-5p and miR-30a-5p expression in WBCs and plasma. We present novel data on the involvement of a possible regulatory axis lncRNAs/miR-30e/CHI3L1 in SSc and hypothesize that MALAT1 and NEAT1 could act as miR-30e-5p and miR-30a-5p decoys. This may be a reason for the increased serum levels of CHI3L1 in SSc patients.
Keywords: CHI3L1; MALAT1; NEAT1; miR-30e; systemic sclerosis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
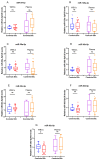
miR-30a-5p and miR-30e-5p are downregulated in SSc. Box plot of (A) miR-24-3p, (C) miR-30a-5p, (D) miR-30b-5p, (E) miR-30c-5p, (G) miR-30e-5p, (F) miR-30d-5p and (B) miR-125a-3p measured in both WBC and plasma of SSc (WBC, n = 31; plasma, n = 30) and healthy controls (WBC, n = 12; plasma, n = 9). Expression levels were determined by TagMan assay after total RNA extraction and presented as fold difference. snoRNA U48 and U6 were used as miRNA normalizers. The data are summarized from two technical replicates for each examined individual. p > 0.05 was marked with *.
Figure 2
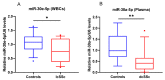
miR-30e-5p and miR-30a-5p are differentially expressed in SSc subgroups. Box plot of miR-30e-5p (A) in WBCs of lcSSc patients (n = 12) and miR-30a-5p (B) in plasma of dcSSc patients (n = 14), compared to healthy controls (WBC, n = 12; plasma, n = 9). Expression levels were determined by TagMan assay after total RNA extraction and presented as fold difference. snoRNA U48 and U6 were used as miRNA normalizers. The data are summarized from two technical replicates for each examined individual. p > 0.05 was marked with *; p > 0.01 was marked with **.
Figure 3
Binding sites of miR-30e-5p (A) and miR-30a-5p (B) in CHI3L1mRNA and MALAT1. DIANA-LncBase v3 and Starbas bioinformatical tools were applied to predict the binding sequence of the two down regulated miRNAs: miR-30e-5p and miR-30a-5p in the 3’UTR of CHI3L1mRNA and lncRNA MALAT1. The in silico analysis revealed that both have a conserved seed region by which they could bind to the complementary sites in CHI3L1mRNA and lncRNA MALAT1 molecules.
Figure 4
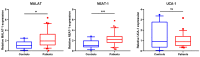
MALAT1 and NEAT1 are upregulated in SSc WBCs. MALAT1 and NEAT1 are induced in WBCs of SSc patients, compared with the control subjects. Expression levels of MALAT1, NEAT1 and UCA1 were evaluated in WBCs of patients (n = 31) and controls (n = 12) by RT-qPCR after total RNA extraction and presented as fold difference. GAPDH, ACTINβ and hUBC were used as RNA normalizers. The data are summarized from two technical replicates for each examined individual. p > 0.05 was marked with *; p > 0.01 was marked with **.
Figure 5
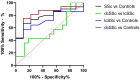
Discriminatory potential of plasma miR-30a-5p levels. ROC analysis was performed to access the discriminatory power of miR-30a-5p in plasma. The AUC values show that miR-30a-5p has the potential as a biomarker for distinguishing SSc from healthy subjects.
Figure 6
Possible lncRNAs/miR30e/CHI3L1regulatory axis in SSc. In SSc, the expression of MALAT1 and NEAT1 in WBCs is upregulated. MALAT1 and NEAT1 can act as competitive endogenous RNAs for miR-30e. Interaction between MALAT1/NEAT1 and miR-30e-5p would abolish the suppressive effect of miR-30e-5p over CHI3L1production, leading to increased serum levels of the glycoprotein in patients with SSc. ↑ indicates induction, whereas ↓ shows downregulation.
Similar articles
- LncRNA NEAT1 and MALAT1 are involved in polycystic ovary syndrome pathogenesis by functioning as competing endogenous RNAs to control the expression of PCOS-related target genes.
ElMonier AA, El-Boghdady NA, Fahim SA, Sabry D, Elsetohy KA, Shaheen AA. ElMonier AA, et al. Noncoding RNA Res. 2023 Mar 3;8(2):263-271. doi: 10.1016/j.ncrna.2023.02.008. eCollection 2023 Jun. Noncoding RNA Res. 2023. PMID: 36935861 Free PMC article. - LncRNA KCNQ1OT1 Promotes Proliferation, Invasion and Metastasis of Prostate Cancer by Regulating miR-211-5p/CHI3L1 Pathway.
Hao H, Chen H, Xie L, Liu H, Wang D. Hao H, et al. Onco Targets Ther. 2021 Mar 3;14:1659-1671. doi: 10.2147/OTT.S288785. eCollection 2021. Onco Targets Ther. 2021. PMID: 33688211 Free PMC article. - Long Non-Coding MALAT1 Functions as a Competing Endogenous RNA to Regulate Vimentin Expression by Sponging miR-30a-5p in Hepatocellular Carcinoma.
Pan Y, Tong S, Cui R, Fan J, Liu C, Lin Y, Tang J, Xie H, Lin P, Zheng T, Yu X. Pan Y, et al. Cell Physiol Biochem. 2018;50(1):108-120. doi: 10.1159/000493962. Epub 2018 Oct 2. Cell Physiol Biochem. 2018. PMID: 30278452 - LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3 axis.
Zhang Y, Wang F, Chen G, He R, Yang L. Zhang Y, et al. Cell Biosci. 2019 Jul 1;9:54. doi: 10.1186/s13578-019-0302-2. eCollection 2019. Cell Biosci. 2019. PMID: 31304004 Free PMC article. Review. - Non-Coding RNAs in the Etiology and Control of Major and Neglected Human Tropical Diseases.
Tamgue O, Mezajou CF, Ngongang NN, Kameni C, Ngum JA, Simo USF, Tatang FJ, Akami M, Ngono AN. Tamgue O, et al. Front Immunol. 2021 Oct 19;12:703936. doi: 10.3389/fimmu.2021.703936. eCollection 2021. Front Immunol. 2021. PMID: 34737736 Free PMC article. Review.
Cited by
- LncRNA NEAT1 and MALAT1 are involved in polycystic ovary syndrome pathogenesis by functioning as competing endogenous RNAs to control the expression of PCOS-related target genes.
ElMonier AA, El-Boghdady NA, Fahim SA, Sabry D, Elsetohy KA, Shaheen AA. ElMonier AA, et al. Noncoding RNA Res. 2023 Mar 3;8(2):263-271. doi: 10.1016/j.ncrna.2023.02.008. eCollection 2023 Jun. Noncoding RNA Res. 2023. PMID: 36935861 Free PMC article. - YKL-40 and the Cellular Metabolic Profile in Parkinson's Disease.
Gevezova M, Kazakova M, Trenova A, Sarafian V. Gevezova M, et al. Int J Mol Sci. 2023 Nov 14;24(22):16297. doi: 10.3390/ijms242216297. Int J Mol Sci. 2023. PMID: 38003487 Free PMC article. - Bioenergetic and Inflammatory Alterations in Regressed and Non-Regressed Patients with Autism Spectrum Disorder.
Gevezova M, Ivanov Z, Pacheva I, Timova E, Kazakova M, Kovacheva E, Ivanov I, Sarafian V. Gevezova M, et al. Int J Mol Sci. 2024 Jul 27;25(15):8211. doi: 10.3390/ijms25158211. Int J Mol Sci. 2024. PMID: 39125780 Free PMC article. - Importance of Studying Non-Coding RNA in Children and Adolescents with Type 1 Diabetes.
Cabiati M, Federico G, Del Ry S. Cabiati M, et al. Biomedicines. 2024 Sep 2;12(9):1988. doi: 10.3390/biomedicines12091988. Biomedicines. 2024. PMID: 39335501 Free PMC article. Review. - [Role and mechanisms of CHI3L1 in coronary artery lesions in a mouse model of Kawasaki disease-like vasculitis].
Cao Y, Gao S, Luo G, Zhao SY, Tang YQ, DU ZH, Pan SL. Cao Y, et al. Zhongguo Dang Dai Er Ke Za Zhi. 2023 Dec 15;25(12):1227-1233. doi: 10.7499/j.issn.1008-8830.2309080. Zhongguo Dang Dai Er Ke Za Zhi. 2023. PMID: 38112139 Free PMC article. Chinese.
References
- Orlandi M., Barsotti S., Lepri G., Codullo V., Di Battista M., Guiducci S., Della Rossa A. One year in review 2018: Systemic sclerosis. Clin. Exp. Rheumatol. 2018;36((Suppl. 113)):3–23. - PubMed
- Dolcino M., Tinazzi E., Puccetti A., Lunardi C. In Systemic Sclerosis, a Unique Long Non Coding RNA Regulates Genes and Pathways Involved in the Three Main Features of the Disease (Vasculopathy, Fibrosis and Autoimmunity) and in Carcinogenesis. J. Clin. Med. 2019;8:320. doi: 10.3390/jcm8030320. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials