Lacticaseibacillus rhamnosus: A Suitable Candidate for the Construction of Novel Bioengineered Probiotic Strains for Targeted Pathogen Control - PubMed (original) (raw)
Review
Lacticaseibacillus rhamnosus: A Suitable Candidate for the Construction of Novel Bioengineered Probiotic Strains for Targeted Pathogen Control
Moloko G Mathipa-Mdakane et al. Foods. 2022.
Abstract
Probiotics, with their associated beneficial effects, have gained popularity for the control of foodborne pathogens. Various sources are explored with the intent to isolate novel robust probiotic strains with a broad range of health benefits due to, among other mechanisms, the production of an array of antimicrobial compounds. One of the shortcomings of these wild-type probiotics is their non-specificity. A pursuit to circumvent this limitation led to the advent of the field of pathobiotechnology. In this discipline, specific pathogen gene(s) are cloned and expressed into a given probiotic to yield a novel pathogen-specific strain. The resultant recombinant probiotic strain will exhibit enhanced species-specific inhibition of the pathogen and its associated infection. Such probiotics are also used as vehicles to deliver therapeutic agents. As fascinating as this approach is, coupled with the availability of numerous probiotics, it brings a challenge with regard to deciding which of the probiotics to use. Nonetheless, it is indisputable that an ideal candidate must fulfil the probiotic selection criteria. This review aims to show how Lacticaseibacillus rhamnosus, a clinically best-studied probiotic, presents as such a candidate. The objective is to spark researchers' interest to conduct further probiotic-engineering studies using L. rhamnosus, with prospects for the successful development of novel probiotic strains with enhanced beneficial attributes.
Keywords: Lacticaseibacillus rhamnosus; pathobiotechnology; pathogen; probiotics; recombinant probiotics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
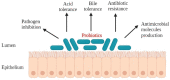
Figure 1
Desirable properties of the probiotic microorganisms.
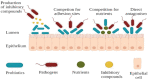
Figure 2
Mechanisms of pathogen inhibition by probiotics.
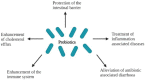
Figure 3
Health effects of probiotics in the host.
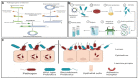
Figure 4
A schematic representation of the steps followed in development of bioengineered probiotics and the mechanism of pathogen inhibition by the bioengineered probiotic strain. (A) Cloning and expression of genes of interest from the pathogen into an expression vector. (B) Transformation of the expression vector carrying the gene of interest from the pathogen into the probiotic and analyses conducted to confirm expression of the foreign gene by the bioengineered probiotic strain. (C) Competitive binding of the bioengineered probiotic strain to the same host receptors to which the target pathogen binds to cause an infection.
Similar articles
- Metabolic Regulation of Longevity and Immune Response in Caenorhabditis elegans by Ingestion of Lacticaseibacillus rhamnosus IDCC 3201 Using Multi-Omics Analysis.
Lee DJ, Eor JY, Kwak MJ, Lee J, Kang AN, Mun D, Choi H, Song M, Kim JN, Kim JM, Yang J, Kim HW, Oh S, Kim Y. Lee DJ, et al. J Microbiol Biotechnol. 2024 May 28;34(5):1109-1118. doi: 10.4014/jmb.2402.02025. Epub 2024 Apr 3. J Microbiol Biotechnol. 2024. PMID: 38563104 Free PMC article. - Enumeration of Probiotic Strain Lacticaseibacillus rhamnosus GG (ATCC 53103) Using Viability Real-time PCR.
Shehata HR, Newmaster SG. Shehata HR, et al. Probiotics Antimicrob Proteins. 2021 Dec;13(6):1611-1620. doi: 10.1007/s12602-021-09849-6. Epub 2021 Sep 30. Probiotics Antimicrob Proteins. 2021. PMID: 34591288 - Probiotic engineering: towards development of robust probiotic strains with enhanced functional properties and for targeted control of enteric pathogens.
Mathipa MG, Thantsha MS. Mathipa MG, et al. Gut Pathog. 2017 May 8;9:28. doi: 10.1186/s13099-017-0178-9. eCollection 2017. Gut Pathog. 2017. PMID: 28491143 Free PMC article. Review. - Bacteraemia Caused by Probiotic Strains of _Lacticaseibacillus rhamnosus_-Case Studies Highlighting the Need for Careful Thought before Using Microbes for Health Benefits.
Mikucka A, Deptuła A, Bogiel T, Chmielarczyk A, Nurczyńska E, Gospodarek-Komkowska E. Mikucka A, et al. Pathogens. 2022 Aug 26;11(9):977. doi: 10.3390/pathogens11090977. Pathogens. 2022. PMID: 36145409 Free PMC article. - Probiotics for the prevention of pediatric antibiotic-associated diarrhea.
Goldenberg JZ, Lytvyn L, Steurich J, Parkin P, Mahant S, Johnston BC. Goldenberg JZ, et al. Cochrane Database Syst Rev. 2015 Dec 22;(12):CD004827. doi: 10.1002/14651858.CD004827.pub4. Cochrane Database Syst Rev. 2015. PMID: 26695080 Updated. Review.
Cited by
- Pharmacotechnical aspects of a stable probiotic formulation toward multidrug-resistance antibacterial activity: design and quality control.
Karimi F, Azadi A, Omidifar N, Najafabady NM, Mohammadi F, Kazemi R, Gholami A. Karimi F, et al. BMC Complement Med Ther. 2023 Oct 31;23(1):391. doi: 10.1186/s12906-023-04224-0. BMC Complement Med Ther. 2023. PMID: 37907893 Free PMC article. - Evaluation of anti-biofilm activity of Lactobacillus rhamnosus GG and Nisin on the expression of _aap, ica-_A and _ica-_D as biofilm-associated genes of Staphylococcus epidermidis.
Dalvand M, Mirhosseini SA, Amini K, Khani S, Mahmoodzadeh Hosseini H, Mansoori K. Dalvand M, et al. Iran J Microbiol. 2023 Aug;15(4):550-556. doi: 10.18502/ijm.v15i4.13509. Iran J Microbiol. 2023. PMID: 38045711 Free PMC article. - Identification and quantification of viable Lacticaseibacillus rhamnosus in probiotics using validated PMA-qPCR method.
Guo L, Ze X, Feng H, Liu Y, Ge Y, Zhao X, Song C, Jiao Y, Liu J, Mu S, Yao S. Guo L, et al. Front Microbiol. 2024 Jan 17;15:1341884. doi: 10.3389/fmicb.2024.1341884. eCollection 2024. Front Microbiol. 2024. PMID: 38298895 Free PMC article. - Oral co-administration of Lactiplantibacillus plantarum 16 and Lacticaseibacillus rhamnosus P118 improves host defense against influenza A virus infection.
Han M, Lu Q, Wang D, Zhou K, Jia C, Teng L, Hamuti A, Peng X, Hu Y, Li W, Yue M, Li Y. Han M, et al. J Virol. 2024 Oct 22;98(10):e0095024. doi: 10.1128/jvi.00950-24. Epub 2024 Sep 11. J Virol. 2024. PMID: 39258911 - Lacticaseibacillus rhamnosus Strain GG (LGG) Regulate Gut Microbial Metabolites, an In Vitro Study Using Three Mature Human Gut Microbial Cultures in a Simulator of Human Intestinal Microbial Ecosystem (SHIME).
Liu L, Narrowe AB, Firrman JA, Mahalak KK, Bobokalonov JT, Lemons JMS, Bittinger K, Daniel S, Tanes C, Mattei L, Friedman ES, Soares JW, Kobori M, Zeng WB, Tomasula PM. Liu L, et al. Foods. 2023 May 24;12(11):2105. doi: 10.3390/foods12112105. Foods. 2023. PMID: 37297350 Free PMC article.
References
- Hill C., Guarner F., Reid G., Gibson G.R., Merenstein D.J., Pot B., Morelli L., Canani R.B., Flint H.J., Salminen S., et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014;11:506–514. doi: 10.1038/nrgastro.2014.66. - DOI - PubMed
- Alshammari E., Patel M., Sachidanandan M., Kumar P., Adnan M. Potential evaluation and health fostering intrinsic traits of novel probiotic strain Enterococcus durans F3 isolated from the gut of fresh water fish Catla catla. Food Sci. Anim. Resour. 2019;39:844–861. doi: 10.5851/kosfa.2019.e57. - DOI - PMC - PubMed
- Mishra S., Acharya S. A brief overview on probiotics: The health friendly microbes. Biomed. Pharmacol. J. 2021;14:1869–2285. doi: 10.13005/bpj/2285. - DOI
- Zheng J., Wittouck S., Salvetti E., Franz C.M., Harris H., Mattarelli P., O’Toole P.W., Pot B., Vandamme P., Walter J., et al. A taxonomic note on the genus Lactobacillus: Description of 23 novel genera, emended description of the genus Lactobacillus Beijerinck 1901, and union of Lactobacillaceae and Leuconostocaceae. Int. J. Syst. Evol. Microbiol. 2020;70:2782–2858. doi: 10.1099/ijsem.0.004107. - DOI - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Research Materials