Loss of Plastid Developmental Genes Coincides With a Reversion to Monoplastidy in Hornworts - PubMed (original) (raw)
Loss of Plastid Developmental Genes Coincides With a Reversion to Monoplastidy in Hornworts
Alexander I MacLeod et al. Front Plant Sci. 2022.
Abstract
The first plastid evolved from an endosymbiotic cyanobacterium in the common ancestor of the Archaeplastida. The transformative steps from cyanobacterium to organelle included the transfer of control over developmental processes, a necessity for the host to orchestrate, for example, the fission of the organelle. The plastids of almost all embryophytes divide independently from nuclear division, leading to cells housing multiple plastids. Hornworts, however, are monoplastidic (or near-monoplastidic), and their photosynthetic organelles are a curious exception among embryophytes for reasons such as the occasional presence of pyrenoids. In this study, we screened genomic and transcriptomic data of eleven hornworts for components of plastid developmental pathways. We found intriguing differences among hornworts and specifically highlight that pathway components involved in regulating plastid development and biogenesis were differentially lost in this group of bryophytes. Our results also confirmed that hornworts underwent significant instances of gene loss, underpinning that the gene content of this group is significantly lower than other bryophytes and tracheophytes. In combination with ancestral state reconstruction, our data suggest that hornworts have reverted back to a monoplastidic phenotype due to the combined loss of two plastid division-associated genes, namely, ARC3 and FtsZ2.
Keywords: bryophytes; hornworts; plant terrestrialization; plastid division; plastid evolution.
Copyright © 2022 MacLeod, Raval, Stockhorst, Knopp, Frangedakis and Gould.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
FIGURE 1
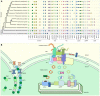
Plastid development and biogenesis in hornworts. (A) A presence/absence pattern (PAP) of various plastid developmental components that are sorted into three categories based on whether they are associated with plastid division (PD) and protein translocation across the plastid envelope via TOC/TIC or the thylakoid membrane. Transparent icons indicate that no gene could be identified. (B) A combined schematic representation of plastid development in embryophytes. Components that are absent from more than two hornworts in our surveyed taxa, or absent in this group altogether, are highlighted by dotted outlines. ARC, accumulation and regulation of chloroplasts; FtsZ, filamentous temperature Z; IMS, intermembrane space; Sec, secretory; SRP, signal recognition particle; Tat; twin arginine translocation; TOC/TIC, translocator of the outer/inner chloroplast membrane; PDV, plastid division. While ARC5 is absent from the Anthoceros agrestis Bonn ecotype, which we included in our OrthoFinder analyses as the representative for this species, our reciprocal best hit pipeline confirmed that it is present in the Oxford ecotype, with its gene ID being AagrOXF_evm.TU.utg000081l.174. A maximum likelihood (ML) tree was constructed via the IQ-TREE version 2.0.3 software (Minh et al., 2020), using an automated selection model, by concatenating single-copy chloroplast and mitochondrial markers from 65 different hornwort species, and three outgroups (Villarreal and Renner, 2012). Said sequences were aligned with MUSCLE in AliView (Edgar, 2004; Laarson, 2014). Gene trees for orthologs listed on the PAP were generated using the PhyML version 3.0 and IQ-TREE version 2.0.3 softwares using automated selection models (Guindon et al., 2010; Lefort et al., 2017). We used the SHOOT framework (Emms and Kelly, 2021) to extract orthologous sequences from across the Archaeplastida for said trees. We analyzed the genomes and transcriptomes of ten hornworts, along with the genomes of Arabidopsis thaliana and Marchantia polymorpha, to determine the presence of various components involved in plastid development (Lamesch et al., 2012; Bowman et al., 2017; Leebens-Mack et al., 2019; Li et al., 2020; Zhang et al., 2020; Supplementary Table 2). These orthology clusters (orthogroups) were identified using the OrthoFinder version 2.5.4 software (Emms and Kelly, 2015, 2019; Supplementary Table 2). To validate orthogroup presence/absence, we checked for reciprocal best hits using DIAMOND (Buchfink et al., 2015). Due to the difficulty in identifying orthologs for the import protein YCF1 in the Archaeplastida (de Vries et al., 2015), we employed a different strategy to identify orthologs for this gene. We extracted established YCF1 sequences from GenBank and UniProt and used them as queries for DIAMOND.
FIGURE 2
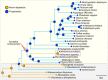
Support for the polyplastidic nature of the ancestral embryophyte and monoplastidic nature of the ancestral hornwort. Pie charts at the nodes display estimates of the probabilities for the plastidic phenotype of the respective most recent common ancestors (MRCAs). Hornworts are highlighted with a white box and a red dotted line. A robust ML species phylogeny of the green lineage was constructed via the IQ-TREE version 2.0.3 (Minh et al., 2020), using an automated selection model, by concatenating several housekeeping genes identified with DIAMOND in 34 different streptophytes, seven chlorophytes, and one glaucophyte (Supplementary Tables 3, 4; Buchfink et al., 2015). We used a reciprocal best hit pipeline with DIAMOND (Buchfink et al., 2015), to analyze the genomes of 34 different streptophytes to determine the presence and absence of orthologs involved in plastid division, to estimate the presence/absence of ARC3 and FtsZ2 at various nodes on our tree (Supplementary Table 5). Subsequent ASRs were undertaken using the ape function from the Phytools package (Revell, 2012).
Similar articles
- The monoplastidic bottleneck in algae and plant evolution.
de Vries J, Gould SB. de Vries J, et al. J Cell Sci. 2018 Jan 29;131(2):jcs203414. doi: 10.1242/jcs.203414. J Cell Sci. 2018. PMID: 28893840 Review. - MONOPLASTIDIC CELL DIVISION IN LOWER LAND PLANTS.
Brown RC, Lemmon BE. Brown RC, et al. Am J Bot. 1990 Apr;77(4):559-571. doi: 10.1002/j.1537-2197.1990.tb13588.x. Am J Bot. 1990. PMID: 30139164 - The quadripolar microtubule system in lower land plants.
Brown RC, Lemmon BE. Brown RC, et al. J Plant Res. 1997 Mar;110(1):93-106. doi: 10.1007/BF02506848. J Plant Res. 1997. PMID: 27520049 - Nuclear genome sequence of the plastid-lacking cryptomonad Goniomonas avonlea provides insights into the evolution of secondary plastids.
Cenci U, Sibbald SJ, Curtis BA, Kamikawa R, Eme L, Moog D, Henrissat B, Maréchal E, Chabi M, Djemiel C, Roger AJ, Kim E, Archibald JM. Cenci U, et al. BMC Biol. 2018 Nov 28;16(1):137. doi: 10.1186/s12915-018-0593-5. BMC Biol. 2018. PMID: 30482201 Free PMC article. - Horizontal and endosymbiotic gene transfer in early plastid evolution.
Ponce-Toledo RI, López-García P, Moreira D. Ponce-Toledo RI, et al. New Phytol. 2019 Oct;224(2):618-624. doi: 10.1111/nph.15965. Epub 2019 Jul 4. New Phytol. 2019. PMID: 31135958 Free PMC article. Review.
Cited by
- A mysterious cloak: the peptidoglycan layer of algal and plant plastids.
MacLeod AI, Knopp MR, Gould SB. MacLeod AI, et al. Protoplasma. 2024 Jan;261(1):173-178. doi: 10.1007/s00709-023-01886-y. Epub 2023 Aug 21. Protoplasma. 2024. PMID: 37603062 Free PMC article. - A molecular atlas of plastid and mitochondrial proteins reveals organellar remodeling during plant evolutionary transitions from algae to angiosperms.
K Raval P, MacLeod AI, Gould SB. K Raval P, et al. PLoS Biol. 2024 May 7;22(5):e3002608. doi: 10.1371/journal.pbio.3002608. eCollection 2024 May. PLoS Biol. 2024. PMID: 38713727 Free PMC article. - What can hornworts teach us?
Frangedakis E, Marron AO, Waller M, Neubauer A, Tse SW, Yue Y, Ruaud S, Waser L, Sakakibara K, Szövényi P. Frangedakis E, et al. Front Plant Sci. 2023 Mar 8;14:1108027. doi: 10.3389/fpls.2023.1108027. eCollection 2023. Front Plant Sci. 2023. PMID: 36968370 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources