The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice - PubMed (original) (raw)
. 2022 Aug;20(8):1470-1486.
doi: 10.1111/pbi.13825. Epub 2022 May 6.
Tian Ma 3, Dingyang Yuan 4 5, Yang Zhou 6, Yan Long 1 2, Ziwen Li 1 2, Zhenying Dong 1 2, Meijuan Duan 5, Dong Yu 5, Yizhi Jing 1, Xiaoyue Bai 1, Yanbo Wang 1, Quancan Hou 1 2, Shuangshuang Liu 1 2, Jin-Song Zhang 6, Shou-Yi Chen 6, Dayong Li 7, Xue Liu 7, Zhikang Li 8, Wensheng Wang 8, Jinping Li 2, Xun Wei 1 2, Biao Ma 3, Xiangyuan Wan 1 2
Affiliations
- PMID: 35403801
- PMCID: PMC9342608
- DOI: 10.1111/pbi.13825
The OsEIL1-OsERF115-target gene regulatory module controls grain size and weight in rice
Chang Liu et al. Plant Biotechnol J. 2022 Aug.
Abstract
Grain size is one of the essential determinants of rice yield. Our previous studies revealed that ethylene plays an important role in grain-size control; however, the precise mechanism remains to be determined. Here, we report that the ethylene response factor OsERF115 functions as a key downstream regulator for ethylene-mediated grain development. OsERF115 encodes an AP2/ERF-type transcriptional factor that is specifically expressed in young spikelets and developing caryopses. Overexpression of OsERF115 significantly increases grain length, width, thickness and weight by promoting longitudinal elongation and transverse division of spikelet hull cells, as well as enhancing grain-filling activity, whereas its knockout mutations lead to the opposite effects, suggesting that OsERF115 positively regulates grain size and weight. OsERF115 transcription is strongly induced by ethylene, and OsEIL1 directly binds to the promoter to activate its expression. OsERF115 acts as a transcriptional repressor to directly or indirectly modulate a set of grain-size genes during spikelet growth and endosperm development. Importantly, haplotype analysis reveals that the SNP variations in the EIN3-binding sites of OsERF115 promoter are significantly associated with the OsERF115 expression levels and grain weight, suggesting that natural variations in the OsERF115 promoter contribute to grain-size diversity. In addition, the OsERF115 orthologues are identified only in grass species, implying a conserved and unique role in the grain development of cereal crops. Our results provide insights into the molecular mechanism of ethylene-mediated grain-size control and a potential strategy based on the OsEIL1-OsERF115-target gene regulatory module for genetic improvement of rice yield.
Keywords: OsEIL1; OsERF115; grain size and weight; regulatory module; rice.
© 2022 The Authors. Plant Biotechnology Journal published by Society for Experimental Biology and The Association of Applied Biologists and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Figure 1
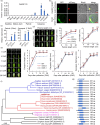
Characterization of OsERF115 gene. (a) RT‐qPCR analysis of OsERF115 expression in various organs of rice plants (Japonica variety Nipponbare). Data are means ± SD, n = 3. (b) Subcellular localization of OsERF115 in rice protoplasts. GHD7‐mCherry plasmid was used as a nuclear marker. Scale bars, 10 μm. (c) RT‐qPCR analysis of OsERF115 expression in its two overexpressing (OX) lines. Data are means ± SD, n = 3. (d, e) Growth analysis of spikelet hulls (d) and developing caryopses (e) in WT and two _OsERF115_‐OX lines. Scale bars, 1 mm. Statistical data are means ± SD, n = 30. (f) Phylogenetic tree of OsERF115 and its orthologues in plant species. Phylogenetic construction was performed using DNAMAN v8 software (Lynnon Biosoft) with the default parameters. (g) Schematic diagram of the protein structures of OsERF115 and its orthologues in plant species corresponding to (f). The AP2/ERF domains are indicated as blue boxes, and the protein lengths are shown on the right. **P < 0.01, ***P < 0.001, Student’s _t_‐test.
Figure 2
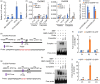
Phenotypic analysis of _OsERF115_‐overexpressing and CRISPR/Cas9 knockout rice lines. (a–c) Morphological comparison of whole plants (a), panicles (b) and grains (c) between WT and two _OsERF115_‐OX lines. Scale bars, 10 cm in ‘a’, 1 cm in ‘b’ and ‘c’. (d) Statistical analysis of grain size and shape in WT and two _OsERF115_‐OX lines. Data are means ± SD, n = 50. (E–K) Yield‐related traits in WT and two _OsERF115_‐OX lines. 1000‐grain weight (e), grain yield per plant (f), plant height (g), tiller number per plant (h), panicle length (i), grain number per plant (j) and seed‐setting rate (k). Data are means ± SD, n = 5‐8. (l) Grain morphology of WT and three OsERF115 knockout lines generated by a CRISPR/Cas9 system. Scale bars, 1 cm. (m) Quantification of grain size and shape of WT and three OsERF115 CRISPR/Cas9 knockout lines. Data are means ± SD, n = 50. (n) 1000‐grain weight of WT and three OsERF115 knockout lines. Data are means ± SD, n = 4. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s _t_‐test. ns, no significance.
Figure 3
OsERF115 promotes longitudinal elongation and transverse division of spikelet hull cells and enhances grain‐filling activity. (a) Scanning electron microscopy (SEM) observation of the glume outer surfaces of WT, OX5‐1 and Cas9‐2 lines mature grains. Scale bar, 2 mm for whole grains and 100 μm for outer glume. (b) Statistical analysis of cell length, width and number in the longitudinal direction in (a). (c) Cross‐sections of spikelet hulls of WT, OX5‐1 and Cas9‐2 lines. Scale bars, 1 mm for whole spikelets (left), 500 μm for cross‐sections (middle) and 100 μm for magnified views (right). Arrows indicate the outer parenchyma layer corresponding to the counted cells. (d) Quantification of cell number and area in the outer parenchyma layer of spikelet hulls. (e) Grain‐filling rate in WT and two _OsERF115‐_OX lines. (f) Expression analysis of grain length‐ and width‐controlling genes in young panicles of WT, OX5‐1 and Cas9‐2 lines by using RT‐qPCR. (g) Expression analysis of grain filling‐related genes in developing caryopses of WT, OX5‐1 and Cas9‐2 lines by using RT‐qPCR. Data are means ± SD, n = 10 (b and d), 20 (e), and 3 (f and g) respectively. *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. ns, no significance.
Figure 4
Ethylene induces OsERF115 expression through OsEIL1‐mediated direct transcriptional activation. (a) Induction of OsERF115 expression by ethylene treatment (100 ppm) in 10 cm panicles of WT and Osein2 mutant as revealed by RT‐qPCR analysis. (b) Expression levels of OsACS1, OsACS4 and OsEIL1 genes in WT young panicles and developing caryopses analysed by RT‐qPCR. (c) Dual‐luciferase assay of OsERF115 promoter activity activated by OsEIL1 or OsEIL2 in rice protoplasts. (d) Four probe positions on OsERF115 promoter and coding region. Probe 2 (P2) and probe 3 (P3) contain the EIN3 binding site (EBS). The EBS sequences in EBS1, EBS2 and EBS3 are shown in red letters. (e) EMSA assay of OsEIL1 binding to the OsERF115 promoter region containing the EBS. (f) The enrichments of OsERF115 promoter analysed by ChIP‐qPCR using the 35S:OsEIL1‐Flag transgenic rice plants. WT plants were used as a negative control. (g) Expression levels of OsERF115 in 10‐cm panicles of WT, Oseil1 mutant and _OsEIL1‐_OX plants analysed by RT‐qPCR. Data are means ± SD, n = 3 (a–c, f and g). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. ns, no significance.
Figure 5
OsERF115 functions as a transcriptional repressor and directly represses OsGW2 and OsGS6 gene expression. (a) Transactivation assay of OsERF115 in rice protoplasts using dual‐luciferase reporter system. Different letters indicate significant differences (P < 0.01, LSD test) in the multiple comparison. (b) Expression levels of OsGW2 and OsGS6 in panicles and caryopses of WT, OX5‐1 and Cas9‐2 lines analysed by RT‐qPCR. (c) Dual‐luciferase assay of promoter activities of OsGW2 and OsGS6 inhibited by TF OsERF115 in rice protoplasts. (d–i) Direct binding of OsERF115 to the promoters of OsGW2 and OsGS6 both in vivo and in vitro, as revealed by EMSA assay (e and h) and ChIP‐qPCR analysis using 35S:OsERF115‐GFP transgenic rice plants (f and i). The 35S:GFP transgenic rice plants were used as a negative control for ChIP‐qPCR analysis. The GCC‐box sequences are shown in red letters in (d) and (g). Data are means ± SD, n = 3 (a–c, f and i). *P < 0.05, **P < 0.01, ***P < 0.001, Student’s t test. ns, no significance.
Figure 6
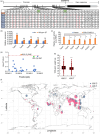
Polymorphisms in OsERF115 promoter are associated with gene expression and grain weight in rice. (a) Haplotype analysis of the OsERF115 gene region in 2486 rice cultivars. AUS aus population, ARO aromatic population, IND indica population, TRJ tropical japonica population, TEJ temperate japonica population. (b) Expression levels of OsERF115 in the five haplotypes with ethylene treatment analysed by RT‐qPCR. Data are means ± SD, n = 3. ***P < 0.001, Student’s t test. ns, not significant. (c) Promoter activity analysis of the OsERF115 promoters with different variations and site‐directed mutations at the two SNPs activated by OsEIL1 in Oseil1 protoplasts. Data are means ± SD, n = 3. The asterisks indicate significant differences compared with Type 1 promoter activated by OsEIL1. *P < 0.05, **P < 0.01, Student’s t test. (d) Association testing between 1000‐grain weight and SNPs in the promoter. The dots marked in orange or green represent the SNPs located at EBSs. (e) Comparisons of 1000‐grain weight sorted by the −835 position alleles. n = 542 (−835C), 1363 (−835T). ***P < 0.001, Student’s _t_‐test. (f) Geographic origin of rice accessions containing the −835 position alleles.
Figure 7
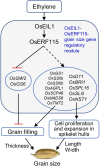
The proposed model for OsERF115 mediating ethylene signalling to regulate grain size in rice. The transcriptional induction of OsERF115 by ethylene depends on OsEIL1 directly activating its expression. OsERF115 indirectly promotes the positive regulatory genes of spikelet hull growth and endosperm development and directly represses OsGW2 and OsGS6 gene expression, thereby promoting grain filling and cell proliferation and expansion of spikelet hulls, ultimately leading to an increase in grain size.
Similar articles
- Molecular Events of Rice AP2/ERF Transcription Factors.
Xie W, Ding C, Hu H, Dong G, Zhang G, Qian Q, Ren D. Xie W, et al. Int J Mol Sci. 2022 Oct 10;23(19):12013. doi: 10.3390/ijms231912013. Int J Mol Sci. 2022. PMID: 36233316 Free PMC article. Review. - MAOHUZI6/ETHYLENE INSENSITIVE3-LIKE1 and ETHYLENE INSENSITIVE3-LIKE2 Regulate Ethylene Response of Roots and Coleoptiles and Negatively Affect Salt Tolerance in Rice.
Yang C, Ma B, He SJ, Xiong Q, Duan KX, Yin CC, Chen H, Lu X, Chen SY, Zhang JS. Yang C, et al. Plant Physiol. 2015 Sep;169(1):148-65. doi: 10.1104/pp.15.00353. Epub 2015 May 20. Plant Physiol. 2015. PMID: 25995326 Free PMC article. - OsEIL1, a rice homolog of the Arabidopsis EIN3 regulates the ethylene response as a positive component.
Mao C, Wang S, Jia Q, Wu P. Mao C, et al. Plant Mol Biol. 2006 May;61(1-2):141-52. doi: 10.1007/s11103-005-6184-1. Plant Mol Biol. 2006. PMID: 16786297 - LARGE GRAIN Encodes a Putative RNA-Binding Protein that Regulates Spikelet Hull Length in Rice.
Chiou WY, Kawamoto T, Himi E, Rikiishi K, Sugimoto M, Hayashi-Tsugane M, Tsugane K, Maekawa M. Chiou WY, et al. Plant Cell Physiol. 2019 Mar 1;60(3):503-515. doi: 10.1093/pcp/pcz014. Plant Cell Physiol. 2019. PMID: 30690508 - Control of grain size in rice.
Li N, Xu R, Duan P, Li Y. Li N, et al. Plant Reprod. 2018 Sep;31(3):237-251. doi: 10.1007/s00497-018-0333-6. Epub 2018 Mar 10. Plant Reprod. 2018. PMID: 29523952 Review.
Cited by
- Genetic Basis of Grain Size and Weight in Rice, Wheat, and Barley.
Gasparis S, Miłoszewski MM. Gasparis S, et al. Int J Mol Sci. 2023 Nov 29;24(23):16921. doi: 10.3390/ijms242316921. Int J Mol Sci. 2023. PMID: 38069243 Free PMC article. Review. - Ethylene in fruits: beyond ripening control.
Huang W, Tan C, Guo H. Huang W, et al. Hortic Res. 2024 Aug 9;11(10):uhae229. doi: 10.1093/hr/uhae229. eCollection 2024 Oct. Hortic Res. 2024. PMID: 39415973 Free PMC article. - The AP2/ERF transcription factor TOE4b regulates photoperiodic flowering and grain yield per plant in soybean.
Li H, Du H, Huang Z, He M, Kong L, Fang C, Chen L, Yang H, Zhang Y, Liu B, Kong F, Zhao X. Li H, et al. Plant Biotechnol J. 2023 Aug;21(8):1682-1694. doi: 10.1111/pbi.14069. Epub 2023 May 12. Plant Biotechnol J. 2023. PMID: 37171033 Free PMC article. - Molecular Events of Rice AP2/ERF Transcription Factors.
Xie W, Ding C, Hu H, Dong G, Zhang G, Qian Q, Ren D. Xie W, et al. Int J Mol Sci. 2022 Oct 10;23(19):12013. doi: 10.3390/ijms231912013. Int J Mol Sci. 2022. PMID: 36233316 Free PMC article. Review. - Multiplex CRISPR-Cas9 knockout of EIL3, EIL4, and EIN2L advances soybean flowering time and pod set.
Cheng Y, Li Y, Yang J, He H, Zhang X, Liu J, Yang X. Cheng Y, et al. BMC Plant Biol. 2023 Oct 27;23(1):519. doi: 10.1186/s12870-023-04543-x. BMC Plant Biol. 2023. PMID: 37884905 Free PMC article.
References
- Azizi, P. , Osman, M. , Hanafi, M.M. , Sahebi, M. , Rafii, M.Y. , Taheri, S. , Harikrishna, J.A. et al. (2019) Molecular insights into the regulation of rice kernel elongation. Crit. Rev. Biotechnol. 39, 904–923. - PubMed
- Bai, X. , Wu, B. and Xing, Y. (2012) Yield‐related QTLs and their applications in rice genetic improvement. J. Integr. Plant Biol. 54, 300–311. - PubMed
- Bradbury, P.J. , Zhang, Z. , Kroon, D.E. , Casstevens, T.M. , Ramdoss, Y. and Buckler, E.S. (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics, 23, 2633–2635. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials