Antimicrobial resistance genes aph(3')-III, erm(B), sul2 and tet(W) abundance in animal faeces, meat, production environments and human faeces in Europe - PubMed (original) (raw)
. 2022 Jun 29;77(7):1883-1893.
doi: 10.1093/jac/dkac133.
Dick J J Heederik 1, Peter Scherpenisse 1, Liese Van Gompel 1, Roosmarijn E C Luiken 1, Katharina Wadepohl 2, Magdalena Skarżyńska 3, Eri Van Heijnsbergen 1, Inge M Wouters 1, Gerdit D Greve 1, Betty G M Jongerius-Gortemaker 1, Monique Tersteeg-Zijderveld 1, Lützen Portengen 1, Katharina Juraschek 4, Jennie Fischer 4, Magdalena Zając 3, Dariusz Wasyl 3, Jaap A Wagenaar 5, Dik J Mevius 5 6, Lidwien A M Smit 1, Heike Schmitt 1 7
Affiliations
- PMID: 35466367
- PMCID: PMC9244224
- DOI: 10.1093/jac/dkac133
Antimicrobial resistance genes aph(3')-III, erm(B), sul2 and tet(W) abundance in animal faeces, meat, production environments and human faeces in Europe
Dongsheng Yang et al. J Antimicrob Chemother. 2022.
Abstract
Background: Real-time quantitative PCR (qPCR) is an affordable method to quantify antimicrobial resistance gene (ARG) targets, allowing comparisons of ARG abundance along animal production chains.
Objectives: We present a comparison of ARG abundance across various animal species, production environments and humans in Europe. AMR variation sources were quantified. The correlation of ARG abundance between qPCR data and previously published metagenomic data was assessed.
Methods: A cross-sectional study was conducted in nine European countries, comprising 9572 samples. qPCR was used to quantify abundance of ARGs [aph(3')-III, erm(B), sul2, tet(W)] and 16S rRNA. Variance component analysis was conducted to explore AMR variation sources. Spearman's rank correlation of ARG abundance values was evaluated between pooled qPCR data and earlier published pooled metagenomic data.
Results: ARG abundance varied strongly among animal species, environments and humans. This variation was dominated by between-farm variation (pigs) or within-farm variation (broilers, veal calves and turkeys). A decrease in ARG abundance along pig and broiler production chains ('farm to fork') was observed. ARG abundance was higher in farmers than in slaughterhouse workers, and lowest in control subjects. ARG abundance showed a high correlation (Spearman's ρ > 0.7) between qPCR data and metagenomic data of pooled samples.
Conclusions: qPCR analysis is a valuable tool to assess ARG abundance in a large collection of livestock-associated samples. The between-country and between-farm variation of ARG abundance could partially be explained by antimicrobial use and farm biosecurity levels. ARG abundance in human faeces was related to livestock antimicrobial resistance exposure.
© The Author(s) 2022. Published by Oxford University Press on behalf of British Society for Antimicrobial Chemotherapy.
Figures
Figure 1.
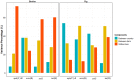
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in all samples. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Asterisk shows the mean by sample type. Pooled faeces, slaughterhouse pig and broiler faeces, and slaughterhouse carcass samples were not included in this figure.
Figure 2.
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in samples related to pig production in the Netherlands. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Only pig farms and slaughterhouses in the Netherlands were involved. Human faeces of control subjects were collected from the ‘Lifelines’ cohort in the Netherlands. Asterisk shows the mean by sample type in the Netherlands.
Figure 3.
Relative abundance of four targets [aph(3′)-III, erm(B), sul2, tet(W)] in samples related to broiler production in Germany. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Only broiler farms and slaughterhouses in Germany were involved. Asterisk shows the mean by sample type in Germany.
Figure 4.
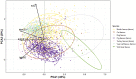
PCA biplot of relative ARG abundance in animal faecal samples. ARG targets: aph(3′)-III, erm(B), sul2, tet(W). Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Symmetric scaling was used. Circles indicate 95% confidence ellipses that were computed with the assumption of multivariate normal distribution of the data.
Figure 5.
Variance component percentages of relative ARG abundance in faecal samples from pigs and broilers. Relative abundance of gene target was calculated by log10 (gene copies/16S copies). Variance percentages of three components (between-country variance, between-farm variance and within-farm variance) were calculated using VCA.
Figure 6.
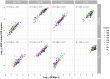
Spearman’s rank correlation of relative ARG abundance and ARG FPKM abundance in pooled faecal samples from pig and broiler farms. ARG targets: aph(3′)-III, erm(B), sul2, tet(W). Relative abundance of gene target was calculated by log10 (gene copies/16S copies). FPKM was log10 transformed after adding a pseudocount of 1. BE, Belgium; BG, Bulgaria; DE, Germany; DK, Denmark; ES, Spain; FR, France; IT, Italy; NL, the Netherlands; PL, Poland.
Similar articles
- Risk factors for the abundance of antimicrobial resistance genes aph(3')-III, erm(B), sul2 and tet(W) in pig and broiler faeces in nine European countries.
Yang D, Heederik DJJ, Mevius DJ, Scherpenisse P, Luiken REC, Van Gompel L, Skarżyńska M, Wadepohl K, Chauvin C, Van Heijnsbergen E, Wouters IM, Greve GD, Jongerius-Gortemaker BGM, Tersteeg-Zijderveld M, Zając M, Wasyl D, Juraschek K, Fischer J, Wagenaar JA, Smit LAM, Schmitt H; EFFORT consortium. Yang D, et al. J Antimicrob Chemother. 2022 Mar 31;77(4):969-978. doi: 10.1093/jac/dkac002. J Antimicrob Chemother. 2022. PMID: 35061866 Free PMC article. - Association of antimicrobial usage with faecal abundance of aph(3')-III, ermB, sul2 and tetW resistance genes in veal calves in three European countries.
Yang D, Van Gompel L, Luiken REC, Sanders P, Joosten P, van Heijnsbergen E, Wouters IM, Scherpenisse P, Chauvin C, Wadepohl K, Greve GD, Jongerius-Gortemaker BGM, Tersteeg-Zijderveld MHG, Soumet C, Skarżyńska M, Juraschek K, Fischer J, Wasyl D, Wagenaar JA, Dewulf J, Schmitt H, Mevius DJ, Heederik DJJ, Smit LAM; EFFORT consortium. Yang D, et al. Int J Antimicrob Agents. 2020 Oct;56(4):106131. doi: 10.1016/j.ijantimicag.2020.106131. Epub 2020 Aug 4. Int J Antimicrob Agents. 2020. PMID: 32763373 - The European livestock resistome.
Munk P, Yang D, Röder T, Maier L, Petersen TN, Duarte ASR, Clausen PTLC, Brinch C, Van Gompel L, Luiken R, Wagenaar JA, Schmitt H, Heederik DJJ, Mevius DJ, Smit LAM; EFFORT Consortium; Bossers A, Aarestrup FM. Munk P, et al. mSystems. 2024 Apr 16;9(4):e0132823. doi: 10.1128/msystems.01328-23. Epub 2024 Mar 19. mSystems. 2024. PMID: 38501800 Free PMC article. - A global baseline for qPCR-determined antimicrobial resistance gene prevalence across environments.
Abramova A, Berendonk TU, Bengtsson-Palme J. Abramova A, et al. Environ Int. 2023 Aug;178:108084. doi: 10.1016/j.envint.2023.108084. Epub 2023 Jul 3. Environ Int. 2023. PMID: 37421899 Review. - Antimicrobial Usage in Animal Production: A Review of the Literature with a Focus on Low- and Middle-Income Countries.
Cuong NV, Padungtod P, Thwaites G, Carrique-Mas JJ. Cuong NV, et al. Antibiotics (Basel). 2018 Aug 15;7(3):75. doi: 10.3390/antibiotics7030075. Antibiotics (Basel). 2018. PMID: 30111750 Free PMC article. Review.
Cited by
- Genotypic Antimicrobial Resistance Profiles of Diarrheagenic Escherichia coli and Nontyphoidal Salmonella Strains Isolated from Children with Diarrhea and Their Exposure Environments in Ethiopia.
Belina D, Gobena T, Kebede A, Chimdessa M, Hald T. Belina D, et al. Infect Drug Resist. 2024 Nov 9;17:4955-4972. doi: 10.2147/IDR.S480395. eCollection 2024. Infect Drug Resist. 2024. PMID: 39539744 Free PMC article. - Recent Trends of Antibiotic Resistance in Staphylococcus aureus Causing Clinical Mastitis in Dairy Herds in Abruzzo and Molise Regions, Italy.
Rossi F, Del Matto I, Saletti MA, Ricchiuti L, Tucci P, Marino L. Rossi F, et al. Antibiotics (Basel). 2023 Feb 21;12(3):430. doi: 10.3390/antibiotics12030430. Antibiotics (Basel). 2023. PMID: 36978297 Free PMC article. - Detection of Acquired Antibiotic Resistance Genes in Domestic Pig (Sus scrofa) and Common Carp (Cyprinus carpio) Intestinal Samples by Metagenomics Analyses in Hungary.
Libisch B, Abdulkadir S, Keresztény T, Papp PP, Olasz F, Fébel H, Sándor ZJ, Rasschaert G, Lambrecht E, Heyndrickx M, Szabó A, Kovács M, Posta K. Libisch B, et al. Antibiotics (Basel). 2022 Oct 20;11(10):1441. doi: 10.3390/antibiotics11101441. Antibiotics (Basel). 2022. PMID: 36290099 Free PMC article. - The Use of Antibiotics and Antimicrobial Resistance in Veterinary Medicine, a Complex Phenomenon: A Narrative Review.
Caneschi A, Bardhi A, Barbarossa A, Zaghini A. Caneschi A, et al. Antibiotics (Basel). 2023 Mar 1;12(3):487. doi: 10.3390/antibiotics12030487. Antibiotics (Basel). 2023. PMID: 36978354 Free PMC article. Review. - City-scale monitoring of antibiotic resistance genes by digital PCR and metagenomics.
Maestre-Carballa L, Navarro-López V, Martinez-Garcia M. Maestre-Carballa L, et al. Environ Microbiome. 2024 Mar 15;19(1):16. doi: 10.1186/s40793-024-00557-6. Environ Microbiome. 2024. PMID: 38491508 Free PMC article.
References
- WHO . Antimicrobial resistance: global report on surveillance. 2014. https://apps.who.int/iris/handle/10665/112642.
- CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Twentieth Edition: M100. 2011.
- CLSI . Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals—Fourth Edition: VET01. 2013.
- Munk P, Knudsen BE, Lukjancenko Oet al. . Abundance and diversity of the faecal resistome in slaughter pigs and broilers in nine European countries. Nat Microbiol 2018; 3: 898–908. - PubMed
- Van Gompel L, Luiken RE, Sarrazin Set al. . The antimicrobial resistome in relation to antimicrobial use and biosecurity in pig farming, a metagenome-wide association study in nine European countries. J Antimicrob Chemother 2019; 74: 865–76. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical