Discovery of adapalene and dihydrotachysterol as antiviral agents for the Omicron variant of SARS-CoV-2 through computational drug repurposing - PubMed (original) (raw)
Discovery of adapalene and dihydrotachysterol as antiviral agents for the Omicron variant of SARS-CoV-2 through computational drug repurposing
Ozkan Fidan et al. Mol Divers. 2023 Feb.
Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been significantly paralyzing the societies, economies and health care systems around the globe. The mutations on the genome of SARS-CoV-2 led to the emergence of new variants, some of which are classified as "variant of concern" due to their increased transmissibility and better viral fitness. The Omicron variant, as the latest variant of concern, dominated the current COVID-19 cases all around the world. Unlike the previous variants of concern, the Omicron variant has 15 mutations on the receptor-binding domain of spike protein and the changes in the key amino acid residues of S protein can enhance the binding ability of the virus to hACE2, resulting in a significant increase in the infectivity of the Omicron variant. Therefore, there is still an urgent need for treatment and prevention of variants of concern, particularly for the Omicron variant. In this study, an in silico drug repurposing was conducted through the molecular docking of 2890 FDA-approved drugs against the mutant S protein of SARS-CoV-2 for Omicron variant. We discovered promising drug candidates for the inhibition of alarming Omicron variant such as quinestrol, adapalene, tamibarotene, and dihydrotachysterol. The stability of ligands complexed with the mutant S protein was confirmed using MD simulations. The lead compounds were further evaluated for their potential use and side effects based on the current literature. Particularly, adapalene, dihydrotachysterol, levocabastine and bexarotene came into prominence due to their non-interference with the normal physiological processes. Therefore, this study suggests that these approved drugs can be considered as drug candidates for further in vitro and in vivo studies to develop new treatment options for the Omicron variant of SARS-CoV-2.
Keywords: Adapalene; Drug repurposing; Omicron variant; SARS-CoV-2; Vitamin D.
© 2022. The Author(s), under exclusive licence to Springer Nature Switzerland AG.
Figures
Fig. 1
Framework for in silico repurposing of existing drugs against viral S protein of the Omicron variant to temper the pathogenicity of viral infection in humans
Fig. 2
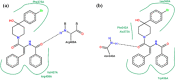
Binding interactions of mutated monomeric subunit of the viral S protein (a) and original non-mutant variant (b) with the reported inhibitor K22
Fig. 3
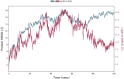
RMSD of the mutated S protein and complexed adapalene recorded during the MD simulation of 100 ns
Fig. 4
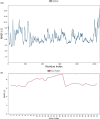
RMSF of the monomeric subunit of the mutated S protein of the Omicron variant (a) and complexed adapalene (b) recorded during the MD simulation for 100 ns
Fig. 5
The detailed contacts observed between macromolecular complex during 100 ns MD simulation. Green-colored bars: hydrogen bonds, blue-colored bars: water bridges, purple-colored bars: hydrophobic interactions, and pink-colored bars: ionic interactions
Similar articles
- Exploring the binding capacity of lactic acid bacteria derived bacteriocins against RBD of SARS-CoV-2 Omicron variant by molecular simulations.
Erol I, Kotil SE, Ortakci F, Durdagi S. Erol I, et al. J Biomol Struct Dyn. 2023 Dec;41(20):10774-10784. doi: 10.1080/07391102.2022.2158934. Epub 2023 Jan 2. J Biomol Struct Dyn. 2023. PMID: 36591650 - Repurposing antibiotics as potent multi-drug candidates for SARS-CoV-2 delta and omicron variants: molecular docking and dynamics.
Serseg T, Linani A, Benarous K, Goumri-Said S. Serseg T, et al. J Biomol Struct Dyn. 2023 Dec;41(20):10377-10387. doi: 10.1080/07391102.2022.2157876. Epub 2022 Dec 21. J Biomol Struct Dyn. 2023. PMID: 36541102 - Omicron: What Makes the Latest SARS-CoV-2 Variant of Concern So Concerning?
Jung C, Kmiec D, Koepke L, Zech F, Jacob T, Sparrer KMJ, Kirchhoff F. Jung C, et al. J Virol. 2022 Mar 23;96(6):e0207721. doi: 10.1128/jvi.02077-21. Epub 2022 Mar 23. J Virol. 2022. PMID: 35225672 Free PMC article. Review. - SARS-CoV-2 Omicron Variant in Medicinal Chemistry Research.
Souza Rocha W, Zhan P, Ferreira da Silva-Júnior E. Souza Rocha W, et al. Curr Top Med Chem. 2023;23(17):1625-1639. doi: 10.2174/1568026623666230411095417. Curr Top Med Chem. 2023. PMID: 37055893 Review.
Cited by
- Discovery of a C-S lyase inhibitor for the prevention of human body malodor formation: tannic acid inhibits the thioalcohol production in Staphylococcus hominis.
Fidan O, Karipcin AD, Köse AH, Anaz A, Demirsoy BN, Arslansoy N, Sun L, Mujwar S. Fidan O, et al. Int Microbiol. 2024 Jun 24. doi: 10.1007/s10123-024-00551-5. Online ahead of print. Int Microbiol. 2024. PMID: 38913231 - Unusual Ni⋯Ni interaction in Ni(ii) complexes as potential inhibitors for the development of new anti-SARS-CoV-2 Omicron drugs.
Singh S, Choudhary M. Singh S, et al. RSC Med Chem. 2024 Feb 20;15(3):895-915. doi: 10.1039/d3md00601h. eCollection 2024 Mar 20. RSC Med Chem. 2024. PMID: 38516589 - Comparative genomics of spike, envelope, and nucleocapsid protein of severe acute respiratory syndrome coronavirus 2.
Khan SS, Ullah A. Khan SS, et al. Afr Health Sci. 2023 Sep;23(3):384-399. doi: 10.4314/ahs.v23i3.45. Afr Health Sci. 2023. PMID: 38357143 Free PMC article. - In silico drug repurposing carvedilol and its metabolites against SARS-CoV-2 infection using molecular docking and molecular dynamic simulation approaches.
Zhang C, Liu J, Sui Y, Liu S, Yang M. Zhang C, et al. Sci Rep. 2023 Dec 4;13(1):21404. doi: 10.1038/s41598-023-48398-6. Sci Rep. 2023. PMID: 38049492 Free PMC article. - Artificial Intelligence in The Management of Neurodegenerative Disorders.
Dhankhar S, Mujwar S, Garg N, Chauhan S, Saini M, Sharma P, Kumar S, Kumar Sharma S, Kamal MA, Rani N. Dhankhar S, et al. CNS Neurol Disord Drug Targets. 2024;23(8):931-940. doi: 10.2174/0118715273266095231009092603. CNS Neurol Disord Drug Targets. 2024. PMID: 37861051 Review.
References
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. - DOI - PMC - PubMed
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. - DOI - PMC - PubMed
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous