The Notable Achievements and the Prospects of Bacterial Pathogen Genomics - PubMed (original) (raw)
The Notable Achievements and the Prospects of Bacterial Pathogen Genomics
Grigorios D Amoutzias et al. Microorganisms. 2022.
Abstract
Throughout the entirety of human history, bacterial pathogens have played an important role and even shaped the fate of civilizations. The application of genomics within the last 27 years has radically changed the way we understand the biology and evolution of these pathogens. In this review, we discuss how the short- (Illumina) and long-read (PacBio, Oxford Nanopore) sequencing technologies have shaped the discipline of bacterial pathogen genomics, in terms of fundamental research (i.e., evolution of pathogenicity), forensics, food safety, and routine clinical microbiology. We have mined and discuss some of the most prominent data/bioinformatics resources such as NCBI pathogens, PATRIC, and Pathogenwatch. Based on this mining, we present some of the most popular sequencing technologies, hybrid approaches, assemblers, and annotation pipelines. A small number of bacterial pathogens are of very high importance, and we also present the wealth of the genomic data for these species (i.e., which ones they are, the number of antimicrobial resistance genes per genome, the number of virulence factors). Finally, we discuss how this discipline will probably be transformed in the near future, especially by transitioning into metagenome-assembled genomes (MAGs), thanks to long-read sequencing.
Keywords: Illumina; Oxford Nanopore; Pacific Biosciences; bacterial pathogen; clinical microbiology; evolution; food-borne pathogens; forensics; genomics; metagenome-assembled genomes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
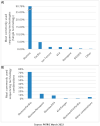
Figure 1
Most frequently used sequencing platforms according to PATRIC, for bacterial pathogens, (A) used as single technology and (B) used in combinations (hybrid approaches).
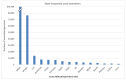
Figure 2
The number of genomes in each bacterial taxonomic group of the NCBI pathogens. (A) The total number of genomes reported in each taxonomic group. (B) The number of complete genomes in each taxonomic group.
Figure 3
Most of the commonly used assemblers reported in the NCBI bacterial pathogens database as of March 2022.
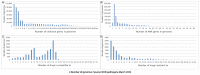
Figure 4
Pathogen annotation data for the virulence, resistance, and drug susceptibility. (A) The number of genomes with a certain number of virulence genes. (B) The number of genomes with a certain number of AMR (antimicrobial resistance) genes. (C) The number of genomes with a certain number of drugs to which they are susceptible (based on experiments). (D) The number of genomes with a certain number of drugs to which they are resistant (based on experiments). Source: The NCBI pathogens (March 2022).
Similar articles
- Completion of draft bacterial genomes by long-read sequencing of synthetic genomic pools.
Derakhshani H, Bernier SP, Marko VA, Surette MG. Derakhshani H, et al. BMC Genomics. 2020 Jul 29;21(1):519. doi: 10.1186/s12864-020-06910-6. BMC Genomics. 2020. PMID: 32727443 Free PMC article. - Lost in plasmids: next generation sequencing and the complex genome of the tick-borne pathogen Borrelia burgdorferi.
Margos G, Hepner S, Mang C, Marosevic D, Reynolds SE, Krebs S, Sing A, Derdakova M, Reiter MA, Fingerle V. Margos G, et al. BMC Genomics. 2017 May 30;18(1):422. doi: 10.1186/s12864-017-3804-5. BMC Genomics. 2017. PMID: 28558786 Free PMC article. - Testing assembly strategies of Francisella tularensis genomes to infer an evolutionary conservation analysis of genomic structures.
Neubert K, Zuchantke E, Leidenfrost RM, Wünschiers R, Grützke J, Malorny B, Brendebach H, Al Dahouk S, Homeier T, Hotzel H, Reinert K, Tomaso H, Busch A. Neubert K, et al. BMC Genomics. 2021 Nov 14;22(1):822. doi: 10.1186/s12864-021-08115-x. BMC Genomics. 2021. PMID: 34773979 Free PMC article. - Microbial Forensics: A Scientific Assessment: This report is based on a colloquium sponsored by the American Academy of Microbiology held June 7-9, 2002, in Burlington, Vermont.
[No authors listed] [No authors listed] Washington (DC): American Society for Microbiology; 2003. Washington (DC): American Society for Microbiology; 2003. PMID: 32809313 Free Books & Documents. Review. - Challenges and opportunities for whole-genome sequencing-based surveillance of antibiotic resistance.
Schürch AC, van Schaik W. Schürch AC, et al. Ann N Y Acad Sci. 2017 Jan;1388(1):108-120. doi: 10.1111/nyas.13310. Ann N Y Acad Sci. 2017. PMID: 28134443 Review.
Cited by
- Economic and health impact modelling of a whole genome sequencing-led intervention strategy for bacterial healthcare-associated infections for England and for the USA.
Fox JM, Saunders NJ, Jerwood SH. Fox JM, et al. Microb Genom. 2023 Aug;9(8):mgen001087. doi: 10.1099/mgen.0.001087. Microb Genom. 2023. PMID: 37555752 Free PMC article. - Antimicrobial Susceptibility and Genetic Prevalence of Extended-Spectrum β-Lactamases in Gram-Negative Rods Isolated from Clinical Specimens in Pakistan.
Idrees MM, Rimsha R, Idrees MD, Saeed A. Idrees MM, et al. Antibiotics (Basel). 2022 Dec 24;12(1):29. doi: 10.3390/antibiotics12010029. Antibiotics (Basel). 2022. PMID: 36671229 Free PMC article. - Genomic classification and antimicrobial resistance profiling of Streptococcus pneumoniae and Haemophilus influenza isolates associated with paediatric otitis media and upper respiratory infection.
Lobb B, Lee MC, McElheny CL, Doi Y, Yahner K, Hoberman A, Martin JM, Hirota JA, Doxey AC, Shaikh N. Lobb B, et al. BMC Infect Dis. 2023 Sep 13;23(1):596. doi: 10.1186/s12879-023-08560-x. BMC Infect Dis. 2023. PMID: 37700242 Free PMC article. - Mining the nanotube-forming Bacillus amyloliquefaciens MR14M3 genome for determining anti-Candida auris and anti-Candida albicans potential by pathogenicity and comparative genomics analysis.
Borgio JF, Alhujaily R, Alquwaie R, Alabdullah MJ, AlHasani E, Alothman W, Alaqeel RK, Alfaraj AS, Kaabi A, Alhur NF, Akhtar S, AlJindan R, Almofty S, Almandil NB, AbdulAzeez S. Borgio JF, et al. Comput Struct Biotechnol J. 2023 Sep 1;21:4261-4276. doi: 10.1016/j.csbj.2023.08.031. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37701018 Free PMC article.
References
- Koch R. Die Ätiologie der Milzbrand-Krankheit, Begründet auf die Entwicklungsgeschichte des Bacillus Anthracis. Robert Koch Institute; Berlin, Germany: 1876. - DOI
Grants and funding
M.N. thanks the University of Thessaly (PhD studentship: DEKA-UTH-259) for their financial support.
LinkOut - more resources
Full Text Sources