A closed Candidatus Odinarchaeum chromosome exposes Asgard archaeal viruses - PubMed (original) (raw)
A closed Candidatus Odinarchaeum chromosome exposes Asgard archaeal viruses
Daniel Tamarit et al. Nat Microbiol. 2022 Jul.
Abstract
Asgard archaea have recently been identified as the closest archaeal relatives of eukaryotes. Their ecology, and particularly their virome, remain enigmatic. We reassembled and closed the chromosome of Candidatus Odinarchaeum yellowstonii LCB_4, through long-range PCR, revealing CRISPR spacers targeting viral contigs. We found related viruses in the genomes of diverse prokaryotes from geothermal environments, including other Asgard archaea. These viruses open research avenues into the ecology and evolution of Asgard archaea.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
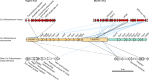
Fig. 1. Ca. Odinarchaeum LCB_4 CRISPR–Cas system and mobile elements.
CRISPR–Cas systems in the Ca. Odinarchaeum LCB_4 chromosome (centre) coloured according to their type classification (orange: I-A; aquamarine: III-D). Full contigs representing mobile elements are shown at the corners, with the vertical lines representing contig boundaries. Viral terminal inverted repeats are represented by hourglass symbols. Connecting lines represent significant full-coverage spacer hits against mobile element targets, shown in black if detected by BlastN (no mismatches: full; one mismatch: dashed) and blue if detected by SpacePHARER and not overlapping with those in black. Source data
Fig. 2. Discovery of additional Asgard archaeal mobile elements.
a, Phylogeny of pPolB obtained with IQTree2 under the Q.pfam+C60+R4+F+PMSF model. Colours: Ca. Odinarchaeum LCB_4 MAG (red); sequences obtained from hot springs (pink); hydrothermal vents (purple); marine water (dark blue); Chatahoochee river (USA) (light blue); mine drainage (brown). Branch support values are FBP (left) and TBE (right). The tree presented is a clade of the full tree shown in Supplementary Fig. 1. b,c, Comparison between the viral contigs of Jordarchaeia QZMA23B3 and Nitrososphaeria SpSt-845 (b) and of Muninn virus and viral contigs in the bins of Ca. Odinarchaeum LCB_4 and Lokiarchaeia E29_bin63 (c). Gene map similarity lines represent reciprocal BlastP hits with an E-value lower than 1 × 10−5 and percentage identity as shown in the upper-right legend.
Extended Data Fig. 1. Obtaining a closed Ca. Odinarchaeum LCB_4 chromosome.
(a) Summary methodology for the reassembly, refinement and closing of the Ca. Odinarchaeum LCB_4 genome. (b) Schematic of the assembly status before long-range PCR (lrPCR), indicating the presence of gaps and the agreement between two separate assemblies, which guided primer design. (c) Purified lrPCR products; lane 1: Invitrogen 1 kb Plus DNA ladder (Thermo Fisher Scientific Inc), 2: Positive control ca. 5 kbp rRNA gene cluster; 3: Positive control ca. 10 kbp ribosomal protein gene cluster; 4-5: first gap closing, at distances of ca. 5 and 5.5 kbp; 6-8: second gap closing, at distances of ca. 4, 4.5 and 5 kbp. Bands of the same sizes were observed 3 times following different cycling parameters, with the clearest visualization shown in this gel. (d) Comparison between previous assembly and new assembly for Huginn virus, indicating circularity. Similarity lines represent two single BlastN hits with up to 1 mismatches. (e) Genomic patterns of the Ca. Odinarchaeum LCB_4 indicating a potential origin of replication at position 959350.
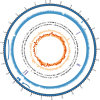
Extended Data Fig. 2. Genome map of Ca. Odinarchaeum LCB_4.
From inside out: (1) GC skew (line) and cumulative GC skew (histogram); (2) GC content; (3) Crick strand genes; (4) Watson strand genes; (5) Nanopore reads coverage capped at 1500X; (6) Illumina read coverage (light: proper pairs, NM < 3) capped at 50X; (7) repeats; (8) chromosome contig.
Extended Data Fig. 3. Predicted structure of selected proteins.
Comparisons between the structures of (a) DJR-MCPs (left: Huginn virus: OLS18934.1; right: Sulfolobus turreted icosahedral virus 1: 3J31); (b) His1-like MCPs (left: Muninn virus: OLS18630.1; right: His1 virus: YP_529533.1); (e) SIRV2-like MCPs (left: Jordarchaeia QZMA23B3: QZMA23B3_25900; right: Sulfolobus islandicus rod-shaped virus 2 (SIRV-2): 3J9X) and (f) transmembrane proteins (left: Muninn virus: OLS18631.1; right: Ca. Odinarchaeum LCB_4 virus: OLS16720). All structures predicted with RoseTTAFold are color-coded according to their error estimate (Å). (c,d) Given the high error estimates for the predicted structures of His1-like MCPs, we append HHsearch results for (C) OLS18630.1 (Muninn virus) and (D) OLS18934.1 (Ca. Odinarchaeum LCB_4 MAG), the latter of which shows a tandem duplication (Regions 1 and 2) of the His1-like MCP. H(h), α-helix; E(e), β-strand; C(c), coil.
Extended Data Fig. 4. Taxonomic placement of archaeal MAGs.
Phylogenomic tree obtained with FastTree including three archaeal MAGs (arrows) containing viral contigs and GTDB Archaea representatives for the phyla Hadarchaeota, Asgard archaea and Thermoproteota. Branch colors within Asgard archaea (orange) represent Jordarchaeia (pink) and Lokiarchaeia (purple). All placements are supported with branch support values of 1.0. Full tree can be found in data repository (see Data Availability statement).
Comment in
- A trove of Asgard archaeal viruses.
Alarcón-Schumacher T, Erdmann S. Alarcón-Schumacher T, et al. Nat Microbiol. 2022 Jul;7(7):931-932. doi: 10.1038/s41564-022-01148-2. Nat Microbiol. 2022. PMID: 35760838 No abstract available.
Similar articles
- Three families of Asgard archaeal viruses identified in metagenome-assembled genomes.
Medvedeva S, Sun J, Yutin N, Koonin EV, Nunoura T, Rinke C, Krupovic M. Medvedeva S, et al. Nat Microbiol. 2022 Jul;7(7):962-973. doi: 10.1038/s41564-022-01144-6. Epub 2022 Jun 27. Nat Microbiol. 2022. PMID: 35760839 Free PMC article. - Exploring the Archaeal Virosphere by Metagenomics.
Zhou Y, Wang Y, Prangishvili D, Krupovic M. Zhou Y, et al. Methods Mol Biol. 2024;2732:1-22. doi: 10.1007/978-1-0716-3515-5_1. Methods Mol Biol. 2024. PMID: 38060114 - Asgard archaeal selenoproteome reveals a roadmap for the archaea-to-eukaryote transition of selenocysteine incorporation machinery.
Huang B, Xiao Y, Zhang Y. Huang B, et al. ISME J. 2024 Jan 8;18(1):wrae111. doi: 10.1093/ismejo/wrae111. ISME J. 2024. PMID: 38896033 Free PMC article. - Asgard archaea: Diversity, function, and evolutionary implications in a range of microbiomes.
MacLeod F, Kindler GS, Wong HL, Chen R, Burns BP. MacLeod F, et al. AIMS Microbiol. 2019 Jan 30;5(1):48-61. doi: 10.3934/microbiol.2019.1.48. eCollection 2019. AIMS Microbiol. 2019. PMID: 31384702 Free PMC article. Review. - Viruses of archaea: Structural, functional, environmental and evolutionary genomics.
Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. Krupovic M, et al. Virus Res. 2018 Jan 15;244:181-193. doi: 10.1016/j.virusres.2017.11.025. Epub 2017 Nov 22. Virus Res. 2018. PMID: 29175107 Free PMC article. Review.
Cited by
- A metagenomic catalogue of the ruminant gut archaeome.
Mi J, Jing X, Ma C, Shi F, Cao Z, Yang X, Yang Y, Kakade A, Wang W, Long R. Mi J, et al. Nat Commun. 2024 Nov 7;15(1):9609. doi: 10.1038/s41467-024-54025-3. Nat Commun. 2024. PMID: 39505912 Free PMC article. - Metagenomic characterization of viruses and mobile genetic elements associated with the DPANN archaeal superphylum.
Wu Z, Liu S, Ni J. Wu Z, et al. Nat Microbiol. 2024 Dec;9(12):3362-3375. doi: 10.1038/s41564-024-01839-y. Epub 2024 Oct 24. Nat Microbiol. 2024. PMID: 39448846 - Complete genomes of Asgard archaea reveal diverse integrated and mobile genetic elements.
Valentin-Alvarado LE, Shi LD, Appler KE, Crits-Christoph A, De Anda V, Adler BA, Cui ML, Ly L, Leão P, Roberts RJ, Sachdeva R, Baker BJ, Savage DF, Banfield JF. Valentin-Alvarado LE, et al. Genome Res. 2024 Oct 29;34(10):1595-1609. doi: 10.1101/gr.279480.124. Genome Res. 2024. PMID: 39406503 Free PMC article. - Asgard archaea modulate potential methanogenesis substrates in wetland soil.
Valentin-Alvarado LE, Appler KE, De Anda V, Schoelmerich MC, West-Roberts J, Kivenson V, Crits-Christoph A, Ly L, Sachdeva R, Greening C, Savage DF, Baker BJ, Banfield JF. Valentin-Alvarado LE, et al. Nat Commun. 2024 Jul 31;15(1):6384. doi: 10.1038/s41467-024-49872-z. Nat Commun. 2024. PMID: 39085194 Free PMC article. - The expanding Asgard archaea invoke novel insights into Tree of Life and eukaryogenesis.
Zhou Z, Liu Y, Anantharaman K, Li M. Zhou Z, et al. mLife. 2022 Dec 18;1(4):374-381. doi: 10.1002/mlf2.12048. eCollection 2022 Dec. mLife. 2022. PMID: 38818484 Free PMC article.
References
Publication types
MeSH terms
Grants and funding
- 817834/ERC_/European Research Council/International
- 203276/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 310039/ERC_/European Research Council/International
- WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources