Viral Circular RNAs and Their Possible Roles in Virus-Host Interaction - PubMed (original) (raw)
Review
Viral Circular RNAs and Their Possible Roles in Virus-Host Interaction
Xing Zhang et al. Front Immunol. 2022.
Abstract
Circular RNAs (circRNAs) as novel regulatory molecules have been recognized in diverse species, including viruses. The virus-derived circRNAs play various roles in the host biological process and the life cycle of the viruses. This review summarized the circRNAs from the DNA and RNA viruses and discussed the biogenesis of viral and host circRNAs, the potential roles of viral circRNAs, and their future perspective. This review will elaborate on new insights gained on viruses encoded circRNAs during virus infection.
Keywords: DNA viruses; RNA viruses; circRNA; host-virus interactions; viral infection.
Copyright © 2022 Zhang, Liang, Wang, Shen, Sun, Gong and Hu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
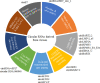
Figure 1
CircRNAs derived from different DNA and RNA viruses with various genome types. MERS-CoV, SARS-CoV-1, SARS-CoV-2, GCRV-873, and BmCPV are RNA viruses. HPV16, HBV, MDV, EBV, KSHV, and MCPyV are DNA viruses. MERS-CoV, SARS-CoV-1, and SARS-CoV-2 belong to Coronaviridae with positive sense and single-strand RNA genome. GCRV-873 and BmCPV belong to Reoarviridae with a double-strand RNA genome. HPV16, HBV, MDV, EBV, KSHV, and MCPyV with double-strand DNA genomes are closely related to the occurrence of tumors or cancers.
Figure 2
Schematic presentation of circRNAs biogenesis and canonical alternative splicing. CircRNAs are classified into six categories according to the formation types. Exonic circRNAs (EcircRNAs), Circular intronic circRNAs (CiRNAs), Exon-intron circRNAs (EIcircRNAs), Fusion circRNAs (F-circRNAs), Read-through circRNAs (Rt-circRNAs), and noncanonical circRNAs like tricRNAs. In canonical alternative splicing, consecutive or selected exons are joined together to generate a series of linear mRNAs to be subsequently translated.
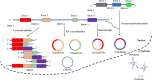
Figure 3
The mechanism of back-splicing and alternative splicing of the pre-mRNA. CircRNA is formed by a 3′,5′ phosphodiester bond using the downstream 5′ splice site (A5SS) and an upstream 3′ splice site (A3SS). Alternative splicing linear RNAs are formed by a 3′,5′ phosphodiester bond using an upstream 5′ splice site (A5SS) and a downstream 3′ splice site (A3SS).
Figure 4
Biological functions of circRNAs. CircRNAs acted as microRNAs/RNA binding protein (RBP) sponges, mediated the alternative splicing and transcription process, regulated the expression levels of parental genes, translated proteins or small peptides, and mRNA trap that the formation of circRNA by back-splicing competes with canonical splicing of linear mRNA.
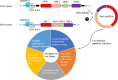
Figure 5
Viral circRNAs produced in the virus’s infected cells may have two-direction regulatory functions mediating the host or virus itself. The DNA viruses with coding regions (ORF) and noncoding regions and RNA viruses without noncoding regions among ORFs were selected to predict the production of viral circRNAs using different biogenesis. These viral circRNAs may have a variety of regulatory roles to host or the virus itself for fitness advantage to viruses.
Similar articles
- Identification and characterization of virus-encoded circular RNAs in host cells.
Chen S, Zheng J, Zhang B, Tang X, Cun Y, Wu T, Xu Y, Ma T, Cheng J, Yu Z, Wang H. Chen S, et al. Microb Genom. 2022 Jun;8(6):mgen000848. doi: 10.1099/mgen.0.000848. Microb Genom. 2022. PMID: 35731570 Free PMC article. - Viruses join the circular RNA world.
Tan KE, Lim YY. Tan KE, et al. FEBS J. 2021 Aug;288(15):4488-4502. doi: 10.1111/febs.15639. Epub 2020 Dec 9. FEBS J. 2021. PMID: 33236482 Free PMC article. Review. - A Novel Intronic Circular RNA Antagonizes Influenza Virus by Absorbing a microRNA That Degrades CREBBP and Accelerating IFN-β Production.
Qu Z, Meng F, Shi J, Deng G, Zeng X, Ge J, Li Y, Liu L, Chen P, Jiang Y, Li C, Chen H. Qu Z, et al. mBio. 2021 Aug 31;12(4):e0101721. doi: 10.1128/mBio.01017-21. Epub 2021 Jul 20. mBio. 2021. PMID: 34281396 Free PMC article. - A Previously Undiscovered Circular RNA, circTNFAIP3, and Its Role in Coronavirus Replication.
Du L, Wang X, Liu J, Li J, Wang S, Lei J, Zhou J, Gu J. Du L, et al. mBio. 2021 Dec 21;12(6):e0298421. doi: 10.1128/mBio.02984-21. Epub 2021 Nov 16. mBio. 2021. PMID: 34781747 Free PMC article. - The role of circular RNAs in viral infection and related diseases.
Xie H, Sun H, Mu R, Li S, Li Y, Yang C, Xu M, Duan X, Chen L. Xie H, et al. Virus Res. 2021 Jan 2;291:198205. doi: 10.1016/j.virusres.2020.198205. Epub 2020 Oct 22. Virus Res. 2021. PMID: 33132144 Free PMC article. Review.
Cited by
- Expression profiles of host miRNAs and circRNAs and ceRNA network during Toxoplasma gondii lytic cycle.
Wang SS, Wang X, He JJ, Zheng WB, Zhu XQ, Elsheikha HM, Zhou CX. Wang SS, et al. Parasitol Res. 2024 Feb 29;123(2):145. doi: 10.1007/s00436-024-08152-x. Parasitol Res. 2024. PMID: 38418741 Free PMC article. - Non-coding RNAs expression in SARS-CoV-2 infection: pathogenesis, clinical significance, and therapeutic targets.
Liu X, Xiong W, Ye M, Lu T, Yuan K, Chang S, Han Y, Wang Y, Lu L, Bao Y. Liu X, et al. Signal Transduct Target Ther. 2023 Dec 6;8(1):441. doi: 10.1038/s41392-023-01669-0. Signal Transduct Target Ther. 2023. PMID: 38057315 Free PMC article. Review. - Regulatory circular RNAs in viral diseases: applications in diagnosis and therapy.
Wang W, Sun L, Huang MT, Quan Y, Jiang T, Miao Z, Zhang Q. Wang W, et al. RNA Biol. 2023 Jan;20(1):847-858. doi: 10.1080/15476286.2023.2272118. Epub 2023 Oct 26. RNA Biol. 2023. PMID: 37882652 Free PMC article. Review. - Functional Involvement of circRNAs in the Innate Immune Responses to Viral Infection.
Maarouf M, Wang L, Wang Y, Rai KR, Chen Y, Fang M, Chen JL. Maarouf M, et al. Viruses. 2023 Aug 5;15(8):1697. doi: 10.3390/v15081697. Viruses. 2023. PMID: 37632040 Free PMC article. Review. - The hidden RNA code: implications of the RNA epitranscriptome in the context of viral infections.
Ribeiro DR, Nunes A, Ribeiro D, Soares AR. Ribeiro DR, et al. Front Genet. 2023 Aug 1;14:1245683. doi: 10.3389/fgene.2023.1245683. eCollection 2023. Front Genet. 2023. PMID: 37614818 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources