Genetic variation associated with condensate dysregulation in disease - PubMed (original) (raw)
. 2022 Jul 25;57(14):1776-1788.e8.
doi: 10.1016/j.devcel.2022.06.010. Epub 2022 Jul 8.
Lena K Afeyan 2, Susana W Hawken 3, Jonathan E Henninger 4, Alessandra Dall'Agnese 4, Victoria E Clark 5, Jesse M Platt 6, Ozgur Oksuz 4, Nancy M Hannett 4, Ido Sagi 4, Tong Ihn Lee 4, Richard A Young 7
Affiliations
- PMID: 35809564
- PMCID: PMC9339523
- DOI: 10.1016/j.devcel.2022.06.010
Genetic variation associated with condensate dysregulation in disease
Salman F Banani et al. Dev Cell. 2022.
Abstract
A multitude of cellular processes involve biomolecular condensates, which has led to the suggestion that diverse pathogenic mutations may dysregulate condensates. Although proof-of-concept studies have identified specific mutations that cause condensate dysregulation, the full scope of the pathological genetic variation that affects condensates is not yet known. Here, we comprehensively map pathogenic mutations to condensate-promoting protein features in putative condensate-forming proteins and find over 36,000 pathogenic mutations that plausibly contribute to condensate dysregulation in over 1,200 Mendelian diseases and 550 cancers. This resource captures mutations presently known to dysregulate condensates, and experimental tests confirm that additional pathological mutations do indeed affect condensate properties in cells. These findings suggest that condensate dysregulation may be a pervasive pathogenic mechanism underlying a broad spectrum of human diseases, provide a strategy to identify proteins and mutations involved in pathologically altered condensates, and serve as a foundation for mechanistic insights into disease and therapeutic hypotheses.
Keywords: Mendelian disease; biomolecular condensates; cancer; clinical genetics; condensate dysregulation; human genetics; pathogenic variants.
Copyright © 2022 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests R.A.Y. is a founder and shareholder of Syros Pharmaceuticals, Camp4 Therapeutics, Omega Therapeutics, and Dewpoint Therapeutics. S.F.B. and A.D. are consultants to Dewpoint Therapeutics. J.E.H. and O.O. are consultants to Camp4 Therapeutics. T.I.L. is a shareholder of Syros Pharmaceuticals and a consultant to Camp4 Therapeutics.
Figures
Figure 1.. A proteome-wide map of pathogenic mutations in condensate-promoting features. See also Figures S1–S2 and Table S1.
A. Multivalent interacting features in proteins that promote biomolecular condensate formation, including modular interacting domains (MIDs, left, green and purple) and low complexity sequences (LCSs, right, blue and green). B. Approach to generate a map of pathogenic mutations that affect condensate-promoting features across the proteome (see also Figure S1A). MIDs and LCSs were mapped across the proteome (left, top) and used to define multivalent proteins (middle). Mendelian and cancer variants were mapped across the proteome (left, bottom), in particular on across the set of multivalent proteins (middle), to identify pathogenic mutations that affect MIDs and LCSs (Methods). The approach allows analysis of diseases, condensates, and mutational signatures associated with pathogenic mutations that affect condensate-promoting features in multivalent proteins (right).
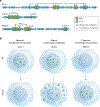
Figure 2.. Condensate dysregulation across the spectrum of disease. See also Table S4I–K.
A. Proportion of pathogenic mutations (depicted as distance from center of radar plot) affecting condensate-promoting features in multivalent proteins across Mendelian diseases. Mendelian diseases are stratified by organ systems in which the diseases had a phenotypic effect (Methods). B. Proportion of pathogenic mutations (depicted as distance from center of radar plot) affecting condensate-promoting features in multivalent proteins across cancers. Cancers are stratified by tissues of origin (Methods). C. Enrichment of GO terms among the set of condensate-forming proteins that have pathogenic mutations that affect condensate-promoting features. GO terms (black dots) are ranked (_x_-axis) by statistical significance (−log10(FDR), _y_-axis). Red line denotes GO term rank corresponding to threshold for statistical significance (FDR < 0.05). The subset of significantly enriched GO terms that correspond to biomolecular condensates (Table S4I) are highlighted (black open circles and labels). Nuclear, cytoplasmic, and plasma membrane-associated condensates are indicated by purple, blue, or gray labels, respectively. D. Significant associations between specific diseases and specific condensates. The set of condensate-forming proteins with pathogenic mutations affecting condensate-promoting features were mapped to specific condensates using Gene Ontology (see Methods) as well as associated with specific diseases. Overlaps between subsets of proteins associated with specific condensates (
y
-axis) and those associated with specific diseases (_x_-axis) were tested for statistical significance. Selected examples of Mendelian diseases (left) and cancer types (right) are shown (see also Table S4J–K). Filled data points correspond to a statistically significant association between the indicated disease with the indicated condensate, with the data point color corresponding to the Benjamini-Hochberg adjusted _p_-value (FDR) for the enrichment of proteins defined as components of the indicated condensate based on GO (Methods) among the set of condensate-forming proteins that have pathogenic mutations involved in the indicated disease that affect condensate-promoting features. Unfilled datapoints correspond to a lack of a statistically significant enrichment. Size of data point is proportional to the fraction of the indicated disease-associated condensate-forming proteins that are components of the indicated condensates.
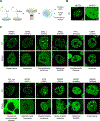
Figure 3.. Pathogenic mutations in condensate-promoting features alter condensate properties in live cells. See also Figures S3–S4 and Table S3.
A. Experimental approach for testing the effect of a subset of identified mutations predicted to affect condensates. N-terminal mEGFP-tagged wild-type or mutant forms of candidate proteins were stably expressed in mESCs and condensate properties were assessed using live cell imaging and quantitative image analysis. B. Representative images of wild-type MECP2 (positive control for condensate incorporation) mEGFP alone (negative control). Nuclei are outlined with white dashed lines. C. Representative images of wild-type versus mutant mEGFP-tagged candidate proteins BARD1, DAXX, SALL1, BRD3, RBM10, BCL11A, NONO, BCOR, TCOF1, HP1A, SRSF2, ESRP1. Specific mutations that were tested along with their associated disease are indicated adjacent to the images.
Figure 4.. Mutations in condensate-promoting features cause diverse condensate dysregulation phenotypes.
Models for observed types of condensate dysregulation resulting from pathogenic mutations that affect condensate-promoting features of condensate-forming proteins, including altered condensate incorporation (left), enhanced condensate formation (middle), and altered condensate localization (right). Candidate where these phenotypes were observed (Figure 3C) are listed.
Comment in
- Charting the human disease condensate dysregulome.
Chandra B, Kriwacki R. Chandra B, et al. Dev Cell. 2022 Jul 25;57(14):1677-1679. doi: 10.1016/j.devcel.2022.07.001. Dev Cell. 2022. PMID: 35901780
Similar articles
- ALS-linked mutations impair UBQLN2 stress-induced biomolecular condensate assembly in cells.
Riley JF, Fioramonti PJ, Rusnock AK, Hehnly H, Castañeda CA. Riley JF, et al. J Neurochem. 2021 Oct;159(1):145-155. doi: 10.1111/jnc.15453. Epub 2021 Aug 20. J Neurochem. 2021. PMID: 34129687 Free PMC article. - Protein Condensate Atlas from predictive models of heteromolecular condensate composition.
Saar KL, Scrutton RM, Bloznelyte K, Morgunov AS, Good LL, Lee AA, Teichmann SA, Knowles TPJ. Saar KL, et al. Nat Commun. 2024 Jul 10;15(1):5418. doi: 10.1038/s41467-024-48496-7. Nat Commun. 2024. PMID: 38987300 Free PMC article. - Hitting the mark: Localization of mRNA and biomolecular condensates in health and disease.
Otis JP, Mowry KL. Otis JP, et al. Wiley Interdiscip Rev RNA. 2023 Nov-Dec;14(6):e1807. doi: 10.1002/wrna.1807. Epub 2023 Jul 2. Wiley Interdiscip Rev RNA. 2023. PMID: 37393916 Free PMC article. Review. - ATP-induced crosslinking of a biomolecular condensate.
Coupe S, Fakhri N. Coupe S, et al. bioRxiv [Preprint]. 2023 Apr 18:2023.04.18.535486. doi: 10.1101/2023.04.18.535486. bioRxiv. 2023. PMID: 37131735 Free PMC article. Updated. Preprint. - Biomolecular condensates in cancer biology.
Suzuki HI, Onimaru K. Suzuki HI, et al. Cancer Sci. 2022 Feb;113(2):382-391. doi: 10.1111/cas.15232. Epub 2021 Dec 14. Cancer Sci. 2022. PMID: 34865286 Free PMC article. Review.
Cited by
- Pervasive mislocalization of pathogenic coding variants underlying human disorders.
Lacoste J, Haghighi M, Haider S, Reno C, Lin ZY, Segal D, Qian WW, Xiong X, Teelucksingh T, Miglietta E, Shafqat-Abbasi H, Ryder PV, Senft R, Cimini BA, Murray RR, Nyirakanani C, Hao T, McClain GG, Roth FP, Calderwood MA, Hill DE, Vidal M, Yi SS, Sahni N, Peng J, Gingras AC, Singh S, Carpenter AE, Taipale M. Lacoste J, et al. Cell. 2024 Nov 14;187(23):6725-6741.e13. doi: 10.1016/j.cell.2024.09.003. Epub 2024 Sep 30. Cell. 2024. PMID: 39353438 - Improved predictions of phase behaviour of intrinsically disordered proteins by tuning the interaction range.
Tesei G, Lindorff-Larsen K. Tesei G, et al. Open Res Eur. 2023 Jan 17;2:94. doi: 10.12688/openreseurope.14967.2. eCollection 2022. Open Res Eur. 2023. PMID: 37645312 Free PMC article. - Evaluation of the determinants for improved pluripotency induction and maintenance by engineered SOX17.
Hu H, Ho DHH, Tan DS, MacCarthy CM, Yu CH, Weng M, Schöler HR, Jauch R. Hu H, et al. Nucleic Acids Res. 2023 Sep 22;51(17):8934-8956. doi: 10.1093/nar/gkad597. Nucleic Acids Res. 2023. PMID: 37607832 Free PMC article. - Precise prediction of phase-separation key residues by machine learning.
Sun J, Qu J, Zhao C, Zhang X, Liu X, Wang J, Wei C, Liu X, Wang M, Zeng P, Tang X, Ling X, Qing L, Jiang S, Chen J, Chen TSR, Kuang Y, Gao J, Zeng X, Huang D, Yuan Y, Fan L, Yu H, Ding J. Sun J, et al. Nat Commun. 2024 Mar 26;15(1):2662. doi: 10.1038/s41467-024-46901-9. Nat Commun. 2024. PMID: 38531854 Free PMC article. - Interplay between membranes and biomolecular condensates in the regulation of membrane-associated cellular processes.
Kim N, Yun H, Lee H, Yoo JY. Kim N, et al. Exp Mol Med. 2024 Nov;56(11):2357-2364. doi: 10.1038/s12276-024-01337-5. Epub 2024 Nov 1. Exp Mol Med. 2024. PMID: 39482532 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- R01 MH104610/MH/NIMH NIH HHS/United States
- F32 CA254216/CA/NCI NIH HHS/United States
- T32 DK007191/DK/NIDDK NIH HHS/United States
- T32 CA251062/CA/NCI NIH HHS/United States
- P01 CA155258/CA/NCI NIH HHS/United States
- T32 GM087237/GM/NIGMS NIH HHS/United States
- R01 GM123511/GM/NIGMS NIH HHS/United States
- F31 CA250171/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources