A data compendium associating the genomes of 12,289 Mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics - PubMed (original) (raw)
A data compendium associating the genomes of 12,289 Mycobacterium tuberculosis isolates with quantitative resistance phenotypes to 13 antibiotics
The CRyPTIC Consortium. PLoS Biol. 2022.
Abstract
The Comprehensive Resistance Prediction for Tuberculosis: an International Consortium (CRyPTIC) presents here a data compendium of 12,289 Mycobacterium tuberculosis global clinical isolates, all of which have undergone whole-genome sequencing and have had their minimum inhibitory concentrations to 13 antitubercular drugs measured in a single assay. It is the largest matched phenotypic and genotypic dataset for M. tuberculosis to date. Here, we provide a summary detailing the breadth of data collected, along with a description of how the isolates were selected, collected, and uniformly processed in CRyPTIC partner laboratories across 23 countries. The compendium contains 6,814 isolates resistant to at least 1 drug, including 2,129 samples that fully satisfy the clinical definitions of rifampicin resistant (RR), multidrug resistant (MDR), pre-extensively drug resistant (pre-XDR), or extensively drug resistant (XDR). The data are enriched for rare resistance-associated variants, and the current limits of genotypic prediction of resistance status (sensitive/resistant) are presented by using a genetic mutation catalogue, along with the presence of suspected resistance-conferring mutations for isolates resistant to the newly introduced drugs bedaquiline, clofazimine, delamanid, and linezolid. Finally, a case study of rifampicin monoresistance demonstrates how this compendium could be used to advance our genetic understanding of rare resistance phenotypes. The data compendium is fully open source and it is hoped that it will facilitate and inspire future research for years to come.
Conflict of interest statement
I have read the journal’s policy and the authors of this manuscript have the following competing interests: E.R. is employed by Public Health England and holds an honorary contract with Imperial College London. I.F.L. is Director of the Scottish Mycobacteria Reference Laboratory. S.N. receives funding from German Center for Infection Research, Excellenz Cluster Precision Medicine in Chronic Inflammation, Leibniz Science Campus Evolutionary Medicine of the LUNG (EvoLUNG)tion EXC 2167. P.S. is a consultant at Genoscreen. T.R. is funded by NIH and DoD and receives salary support from the non-profit organization FIND. T.R. is a co-founder, board member and shareholder of Verus Diagnostics Inc, a company that was founded with the intent of developing diagnostic assays. Verus Diagnostics was not involved in any way with data collection, analysis or publication of the results. T.R. has not received any financial support from Verus Diagnostics. UCSD Conflict of Interest office has reviewed and approved T.R.’s role in Verus Diagnostics Inc. T.R. is a co-inventor of a provisional patent for a TB diagnostic assay (provisional patent #: 63/048.989). T.R. is a co-inventor on a patent associated with the processing of TB sequencing data (European Patent Application No. 14840432.0 & USSN 14/912,918). T.R. has agreed to “donate all present and future interest in and rights to royalties from this patent” to UCSD to ensure that he does not receive any financial benefits from this patent. S.S. is working and holding ESOPs at HaystackAnalytics Pvt. Ltd. (Product: Using whole genome sequencing for drug susceptibility testing for Mycobacterium tuberculosis). G.F.G. is listed as an inventor on patent applications for RBD-dimer-based CoV vaccines. The patents for RBD-dimers as protein subunit vaccines for SARS-CoV-2 have been licensed to Anhui Zhifei Longcom Biopharmaceutical Co. Ltd, China.
Figures
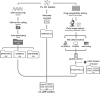
Fig 1. Processing sequencing and minimum inhibitory concentration data for 15,211 Mycobacterium tuberculosis isolates (“full” dataset).
Briefly: Each isolate was DNA sequenced using an Illumina machine and plated onto 96-well plates (UKMYC5/6) containing 5–10× doubling dilutions of 13 antitubercular drugs for DST. Associated metadata (including country of origin and processing laboratory) was recorded. DNA variant calling and analysis was performed using Clockwork and Minos [47]). After 14 days, MIC measurements were taken by a trained scientist using Vizion, and the plate was photographed to also measure the MIC using the automated AMyGDA software and citizen scientists from BashTheBug [45]. After quality control procedures, phenotypic MIC data for 2,922 isolates were removed. The compendium therefore contains 15,211 isolates with WGS data (“full dataset”), 12,289 of which have matched quality assessed phenotypic data (“data compendium”). The raw sequence, VCFs, MICs, and binary resistance calls for the data compendium are presented in “CRyPTIC_reuse_table_20211019.csv” via an FTP site (see Methods), and the raw sequence and VCF files for those samples present in the full dataset are presented in “CRyPTIC_excluded_samples_20220607.tsv” via the same FTP site (see Methods). The data tables GENOTYPES.csv, VARIANTS.csv, MUTATIONS.csv, SAMPLES.csv, and PHENOTYPES.csv used for the analyses presented in this manuscript are also accessible via the FTP site (see Methods). DST, drug susceptibility testing; MIC, minimum inhibitory concentration; WGS, whole-genome sequencing.
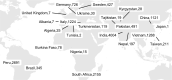
Fig 2. Geographical distribution of 15,211 CRyPTIC Mycobacterium tuberculosis clinical isolates (“full” dataset).
The total number of isolates contributed by each country is depicted. Where the origin of an isolate was not known, the collection site identity was assigned (this occurred for 269 isolates in Germany, 17 isolates in India, 6 isolates in Peru, 885 isolates in Italy, 510 isolates in South Africa, 357 isolates in Sweden, 208 isolates in Taiwan, 1 isolate in Brazil, and 4 isolates in the UK). The base layer map was sourced from Natural Earth, which is in the public domain (see
http://www.naturalearthdata.com/about/terms-of-use/
), and created using the R packages “ggmap” and “maps” (see
github.com/kerrimalone/Brankin_Malone_2022
/).
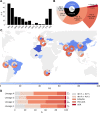
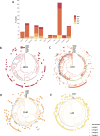
Fig 3. Drug phenotype data for the CRyPTIC compendium.
(A) Frequency of resistance to each of 13 drugs in the data compendium. The total number of isolates with a binary phenotype (of any quality) for the corresponding drug is presented in Table 1. (B) Phenotypes of the 12,289 isolates with a binary phenotype for at least 1 drug. (C) Geographical distribution of phenotypes of 12,289 compendium isolates. Intensity of blue shows the percentage of isolates contributed that were categorised as susceptible to all 13 drugs (“%S”). Donut plots show the proportions of resistant phenotypes identified in (B) for countries contributing > = 100 isolates with drug resistance. (D) Proportions of resistance phenotypes in the 4 major Mycobacterium tuberculosis lineages. N is the number of isolates of the lineage called resistant to at least one of the 13 drugs. The base layer map in (C) was sourced from Natural Earth, which is in the public domain (see
http://www.naturalearthdata.com/about/terms-of-use/
), and created using the python library “geopandas” (see
github.com/kerrimalone/Brankin_Malone_2022
/). AMI, amikacin; BDQ, bedaquiline; CFZ, clofazimine; CRyPTIC, Comprehensive Resistance Prediction for Tuberculosis: an International Consortium; DLM, delamanid; EMB, ethambutol; ETH, ethionamide; INH, isoniazid; KAN, kanamycin; LEV, levofloxacin; LZD, linezolid; MDR, multidrug resistant; MXF, moxifloxacin; pre-XDR, pre-extensively drug resistant; RFB, rifabutin; RIF, rifampicin; RR, rifampicin resistant; XDR, extensively drug resistant.
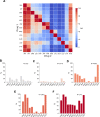
Fig 4. Co-occurrence of resistance to 1 drug conditional on resistance to another drug, or to resistance background.
(A) The heatmap shows the probability of an isolate being resistant to Drug 2 if it is resistant to Drug 1, percentages are given in Table F in S1 File. (B-F) Percentage of isolates that are resistant to another of the 13 drugs in a background of (B) isoniazid susceptible + rifampicin susceptible (but resistant to at least one other antitubercular drug), (C) isoniazid resistant + rifampicin susceptible, (D) MDR/RR, (E) Pre-XDR, and (F) XDR. Only samples with definite phenotypes for RIF in MDR backgrounds and RIF and INH in non-MDR backgrounds and the additional drug are included. AMI, amikacin; BDQ, bedaquiline; CFZ, clofazimine; CRyPTIC, Comprehensive Resistance Prediction for Tuberculosis: an International Consortium; DLM, delamanid; EMB, ethambutol; ETH, ethionamide; INH, isoniazid; KAN, kanamycin; LEV, levofloxacin; LZD, linezolid; MDR, multidrug resistant (resistant to first-line drugs isoniazid and rifampicin); MXF, moxifloxacin; pre-XDR, pre-extensively drug resistant (MDR/RR + fluoroquinolone resistant); RFB, rifabutin; RIF, rifampicin; RR, rifampicin resistant; XDR, extensively drug resistant (MDR/RR + resistant to at least 1 fluoroquinolone and either bedaquiline or linezolid).
Fig 5. Resistance to bedaquiline, clofazimine, delamanid, and linezolid among Mycobacterium tuberculosis compendium isolates.
(A) The prevalence (within these data) of resistance to BDQ, CFZ, DLM, and LZD per country or origin or collection site. Phylotrees are shown for isolates phenotypically resistant to (B) BDQ, (C) CFZ, (D) DLM, and (E) LZD. Tip point colours denote lineage. Each outer track represents a gene thought to be associated with resistance and coloured blocks denote the presence of a nonsynonymous mutation in the relevant gene for a given isolate. Mutations in these genes that are either associated with sensitivity or present in >5% of the collection of isolates as a whole were ignored. BDQ, bedaquiline; CFZ, clofazimine; DLM, delamanid; LZD, linezolid.
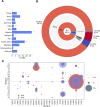
Fig 6. Rifampicin monoresistance.
(A) Percentage of RR isolates that are RMR by country of isolate origin. * indicates RMR proportions that were significantly different from that of the total dataset using a 2-tailed z-test with 95% confidence. (B) MDR predictions for RR isolates made using the Xpert MTB/RIF assay proxy. N is the total number of RR isolates. The inner ring shows the proportion of RR isolates that are RMR and MDR. The middle ring represents the proportions of RMR and MDR isolates that have an SNP (synonymous or nonsynonymous) in the RRDR of rpoB (“RRDR”), no RRDR SNP but a SNP elsewhere in the rpoB gene (“_rpoB_”), and no rpoB mutations (“none”). The outer ring shows the expected TP, TN, FP, and FN MDR predictions of Xpert MTB/RIF assay, based on the SNPs present in the RR isolates. (C) Nonsynonymous mutations found in the RRDR of rpoB in RMR isolates and MDR isolates. Presence of a coloured spot indicates that the mutation was found in RMR/MDR isolates, and spot size corresponds to the proportion of RMR or MDR isolates carrying that mutation. FN, false negative; FP, false positive; MDR, multidrug resistant; RRDR, rifampicin resistance–determining region; RMR, rifampicin monoresistant; RR, rifampicin resistant; SNP, single nucleotide polymorphism; TN, true negative; TP, true positive.
Similar articles
- Whole-genome sequencing of clinical isolates from tuberculosis patients in India: real-world data indicates a high proportion of pre-XDR cases.
Bhanushali A, Atre S, Nair P, Thandaseery GA, Shah S, Kuruwa S, Zade A, Nikam C, Gomare M, Chatterjee A. Bhanushali A, et al. Microbiol Spectr. 2024 May 2;12(5):e0277023. doi: 10.1128/spectrum.02770-23. Epub 2024 Apr 10. Microbiol Spectr. 2024. PMID: 38597637 Free PMC article. - High clustering rate and genotypic drug-susceptibility screening for the newly recommended anti-tuberculosis drugs among global extensively drug-resistant Mycobacterium tuberculosis isolates.
Trisakul K, Nonghanphithak D, Chaiyachat P, Kaewprasert O, Sakmongkoljit K, Reechaipichitkul W, Chaiprasert A, Blair D, Clark TG, Faksri K. Trisakul K, et al. Emerg Microbes Infect. 2022 Dec;11(1):1857-1866. doi: 10.1080/22221751.2022.2099304. Emerg Microbes Infect. 2022. PMID: 35792049 Free PMC article. - First report of whole-genome analysis of an extensively drug-resistant Mycobacterium tuberculosis clinical isolate with bedaquiline, linezolid and clofazimine resistance from Uganda.
Kabahita JM, Kabugo J, Kakooza F, Adam I, Guido O, Byabajungu H, Namutebi J, Namaganda MM, Lutaaya P, Otim J, Kakembo FE, Kanyerezi S, Nabisubi P, Sserwadda I, Kasule GW, Nakato H, Musisi K, Oola D, Joloba ML, Mboowa G. Kabahita JM, et al. Antimicrob Resist Infect Control. 2022 May 12;11(1):68. doi: 10.1186/s13756-022-01101-2. Antimicrob Resist Infect Control. 2022. PMID: 35550202 Free PMC article. - A Narrative Review of Bedaquiline and Delamanid: New Arsenals Against Multidrug-Resistant and Extensively Drug-Resistant Mycobacterium tuberculosis.
Ahmad Khosravi N, Sirous M, Khosravi AD, Saki M. Ahmad Khosravi N, et al. J Clin Lab Anal. 2024 Aug;38(15-16):e25091. doi: 10.1002/jcla.25091. J Clin Lab Anal. 2024. PMID: 39431709 Free PMC article. Review. - Mechanisms of drug resistance in Mycobacterium tuberculosis.
Zhang Y, Yew WW. Zhang Y, et al. Int J Tuberc Lung Dis. 2009 Nov;13(11):1320-30. Int J Tuberc Lung Dis. 2009. PMID: 19861002 Review.
Cited by
- Evolution of Mycobacterium tuberculosis drug resistance in the genomic era.
Nimmo C, Millard J, Faulkner V, Monteserin J, Pugh H, Johnson EO. Nimmo C, et al. Front Cell Infect Microbiol. 2022 Oct 7;12:954074. doi: 10.3389/fcimb.2022.954074. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36275027 Free PMC article. Review. - Genomic epidemiology analysis of drug-resistant Mycobacterium tuberculosis distributed in Mexico.
Mejía-Ponce PM, Ramos-González EJ, Ramos-García AA, Lara-Ramírez EE, Soriano-Herrera AR, Medellín-Luna MF, Valdez-Salazar F, Castro-Garay CY, Núñez-Contreras JJ, De Donato-Capote M, Sharma A, Castañeda-Delgado JE, Zenteno-Cuevas R, Enciso-Moreno JA, Licona-Cassani C. Mejía-Ponce PM, et al. PLoS One. 2023 Oct 13;18(10):e0292965. doi: 10.1371/journal.pone.0292965. eCollection 2023. PLoS One. 2023. PMID: 37831695 Free PMC article. - Genomic characterization of multidrug-resistant tuberculosis in Shanghai, China: antibiotic resistance, virulence and transmission.
Guo Y, Lu J, Jin P, Qiu Z, Yu F, Zhu Y, Huang J. Guo Y, et al. JAC Antimicrob Resist. 2025 May 8;7(3):dlaf064. doi: 10.1093/jacamr/dlaf064. eCollection 2025 Jun. JAC Antimicrob Resist. 2025. PMID: 40342723 Free PMC article. - The antimicrobial resistance crisis needs action now.
Pariente N; PLOS Biology Staff Editors. Pariente N, et al. PLoS Biol. 2022 Nov 23;20(11):e3001918. doi: 10.1371/journal.pbio.3001918. eCollection 2022 Nov. PLoS Biol. 2022. PMID: 36417490 Free PMC article. - Integrative analysis of multimodal patient data identifies personalized predictors of tuberculosis treatment prognosis.
Sambarey A, Smith K, Chung C, Arora HS, Yang Z, Agarwal PP, Chandrasekaran S. Sambarey A, et al. iScience. 2024 Jan 29;27(2):109025. doi: 10.1016/j.isci.2024.109025. eCollection 2024 Feb 16. iScience. 2024. PMID: 38357663 Free PMC article.
References
- WHO. Global Tuberculosis Report 2020. 2021.
- World Health Organisation. EndTB Campaign. Available from: www.who.int/teams/global-tuberculosis-programme/the-end-tb-strategy.
- Boehme CC, Nicol MP, Nabeta P, Michael JS, Gotuzzo E, Tahirli R, et al.. Feasibility, diagnostic accuracy, and effectiveness of decentralised use of the Xpert MTB/RIF test for diagnosis of tuberculosis and multidrug resistance: a multicentre implementation study. Lancet. 2011;377:1495–1505. doi: 10.1016/S0140-6736(11)60438-8 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U60 OE000103/OE/OSELS CDC HHS/United States
- 214560/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- T32 GM007365/GM/NIGMS NIH HHS/United States
- 203135/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 098316 /WT_/Wellcome Trust/United Kingdom
- 203141/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI124413/AI/NIAID NIH HHS/United States
- MRC_/Medical Research Council/United Kingdom
- 206724/Z/17/Z/WT_/Wellcome Trust/United Kingdom
- R01 AI146338/AI/NIAID NIH HHS/United States
- 201470/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- 101237/Z/13/B/WT_/Wellcome Trust/United Kingdom
- 214321/Z/18/Z/WT_/Wellcome Trust/United Kingdom
- 200205/Z/15/Z/WT_/Wellcome Trust/United Kingdom
- 203583/Z/16/Z/WT_/Wellcome Trust/United Kingdom
- BRC-1215-20008/DH_/Department of Health/United Kingdom
- FC0010218/WT_/Wellcome Trust/United Kingdom
- R01 AI114900/AI/NIAID NIH HHS/United States
- R03 AI117312/AI/NIAID NIH HHS/United States
- FC0010218/CRUK_/Cancer Research UK/United Kingdom
LinkOut - more resources
Full Text Sources