Microbiome-based interventions to modulate gut ecology and the immune system - PubMed (original) (raw)
Review
Microbiome-based interventions to modulate gut ecology and the immune system
Thomas C A Hitch et al. Mucosal Immunol. 2022 Jun.
Abstract
The gut microbiome lies at the intersection between the environment and the host, with the ability to modify host responses to disease-relevant exposures and stimuli. This is evident in how enteric microbes interact with the immune system, e.g., supporting immune maturation in early life, affecting drug efficacy via modulation of immune responses, or influencing development of immune cell populations and their mediators. Many factors modulate gut ecosystem dynamics during daily life and we are just beginning to realise the therapeutic and prophylactic potential of microbiome-based interventions. These approaches vary in application, goal, and mechanisms of action. Some modify the entire community, such as nutritional approaches or faecal microbiota transplantation, while others, such as phage therapy, probiotics, and prebiotics, target specific taxa or strains. In this review, we assessed the experimental evidence for microbiome-based interventions, with a particular focus on their clinical relevance, ecological effects, and modulation of the immune system.
© 2022. The Author(s).
Conflict of interest statement
T.C.A.H. and T.C. have ongoing scientific collaborations with HiPP GmbH. T.C. is also a member of the scientific advisory board of Savanna Ingredients GmbH.
Figures
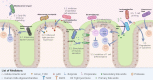
Fig. 1. Constituents and mechanisms of microbiome-based interventions.
a Common constituents of microbiome-based intervention methods are broadly illustrated. b Schematic of the three major categories by which interventions influence the gut microbiome. The simplified native microbiome members are coloured shades of red to purple, while introduced species are shades of blue and prebiotic fibres are green. FMT faecal microbiota transplant, FFT faecal filtrate transplant.
Fig. 2. Visualisation of some discussed mechanisms of how microbiome-based interventions can modulate the immune system.
Each interaction is numbered based on the sub-section they are detailed in. The key mediators and molecules involved are detailed below the scheme. Species known to conduct these interactions are given as examples in black. The mucus layer is depicted in green. As a schematic, the sizes and location of mediators and microbes have been modified to enhance visualisation.
Comment in
- Exposure to environmental microbiota may modulate gut microbial ecology and the immune system.
Stanhope J, Weinstein P. Stanhope J, et al. Mucosal Immunol. 2023 Apr;16(2):99-103. doi: 10.1016/j.mucimm.2023.03.001. Epub 2023 Mar 10. Mucosal Immunol. 2023. PMID: 36906178 No abstract available.
Similar articles
- Microbiomes in physiology: insights into 21st-century global medical challenges.
Shehata E, Parker A, Suzuki T, Swann JR, Suez J, Kroon PA, Day-Walsh P. Shehata E, et al. Exp Physiol. 2022 Apr;107(4):257-264. doi: 10.1113/EP090226. Epub 2022 Feb 14. Exp Physiol. 2022. PMID: 35081663 Free PMC article. Review. - The triple interactions between gut microbiota, mycobiota and host immunity.
Yang J, Yang H, Li Y. Yang J, et al. Crit Rev Food Sci Nutr. 2023 Nov;63(33):11604-11624. doi: 10.1080/10408398.2022.2094888. Epub 2022 Jul 1. Crit Rev Food Sci Nutr. 2023. PMID: 35776086 - Modulating the Microbiome for Crohn's Disease Treatment.
Gowen R, Gamal A, Di Martino L, McCormick TS, Ghannoum MA. Gowen R, et al. Gastroenterology. 2023 Apr;164(5):828-840. doi: 10.1053/j.gastro.2023.01.017. Epub 2023 Jan 24. Gastroenterology. 2023. PMID: 36702360 Free PMC article. Review. - Gastrointestinal symptoms, gut microbiome, probiotics and prebiotics in anorexia nervosa: A review of mechanistic rationale and clinical evidence.
Dhopatkar N, Keeler JL, Mutwalli H, Whelan K, Treasure J, Himmerich H. Dhopatkar N, et al. Psychoneuroendocrinology. 2023 Jan;147:105959. doi: 10.1016/j.psyneuen.2022.105959. Epub 2022 Oct 21. Psychoneuroendocrinology. 2023. PMID: 36327759 Review. - Harnessing the power within: engineering the microbiome for enhanced gynecologic health.
Brennan C, Chan K, Kumar T, Maissy E, Brubaker L, Dothard MI, Gilbert JA, Gilbert KE, Lewis AL, Thackray VG, Zarrinpar A, Knight R. Brennan C, et al. Reprod Fertil. 2024 Apr 15;5(2):e230060. doi: 10.1530/RAF-23-0060. Print 2024 Apr 1. Reprod Fertil. 2024. PMID: 38513356 Free PMC article. Review.
Cited by
- The causal relationship between gut microbiota and constipation: a two-sample Mendelian randomization study.
He N, Sheng K, Li G, Zhang S. He N, et al. BMC Gastroenterol. 2024 Aug 19;24(1):271. doi: 10.1186/s12876-024-03306-8. BMC Gastroenterol. 2024. PMID: 39160466 Free PMC article. - Editorial: Targeting the microbiota to attenuate chronic inflammation.
Moreno E, Trøseid M, Vujkovic-Cvijin I, Marchetti G, Martín-Pedraza L, Serrano-Villar S. Moreno E, et al. Front Immunol. 2023 Apr 18;14:1202222. doi: 10.3389/fimmu.2023.1202222. eCollection 2023. Front Immunol. 2023. PMID: 37153550 Free PMC article. No abstract available. - Mechanistic insights into the role of probiotics in modulating immune cells in ulcerative colitis.
Guo N, Lv LL. Guo N, et al. Immun Inflamm Dis. 2023 Oct;11(10):e1045. doi: 10.1002/iid3.1045. Immun Inflamm Dis. 2023. PMID: 37904683 Free PMC article. Review. - Diagnosing and engineering gut microbiomes.
Cappio Barazzone E, Diard M, Hug I, Larsson L, Slack E. Cappio Barazzone E, et al. EMBO Mol Med. 2024 Nov;16(11):2660-2677. doi: 10.1038/s44321-024-00149-4. Epub 2024 Oct 28. EMBO Mol Med. 2024. PMID: 39468301 Free PMC article. Review. - Targeting nonalcoholic fatty liver disease via gut microbiome-centered therapies.
Koning M, Herrema H, Nieuwdorp M, Meijnikman AS. Koning M, et al. Gut Microbes. 2023 Jan-Dec;15(1):2226922. doi: 10.1080/19490976.2023.2226922. Gut Microbes. 2023. PMID: 37610978 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/F/00044409/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10353/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BBS/E/F/000PR10356/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources