Positive selection-driven fixation of a hominin-specific amino acid mutation related to dephosphorylation in IRF9 - PubMed (original) (raw)
Positive selection-driven fixation of a hominin-specific amino acid mutation related to dephosphorylation in IRF9
Jianhai Chen et al. BMC Ecol Evol. 2022.
Abstract
The arms race between humans and pathogens drives the evolution of the human genome. It is thus expected that genes from the interferon-regulatory factors family (IRFs), a critical family for anti-viral immune response, should be undergoing episodes of positive selection. Herein, we tested this hypothesis and found multiple lines of evidence for positive selection on the amino acid site Val129 (NP_006075.3:p.Ser129Val) of human IRF9. Interestingly, the ancestral reconstruction and population distribution analyses revealed that the ancestral state (Ser129) is conserved among mammals, while the derived positively selected state (Val129) was fixed before the "out-of-Africa" event ~ 500,000 years ago. The motif analysis revealed that this young amino acid (Val129) may serve as a dephosphorylation site of IRF9. Structural parallelism between homologous genes further suggested the functional effects underlying the dephosphorylation that may affect the immune activity of IRF9. This study provides a model in which a strong positive Darwinian selection drives a recent fixation of a hominin-specific amino acid leading to molecular adaptation involving dephosphorylation in an immune-responsive gene.
Keywords: IRF9; Molecular adaptation; Positive selection; Protein phosphorylation; Purifying selection.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing financial interests.
Figures
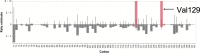
Fig. 1
The FEL method identified positive selection on Val129 (shown with black arrow). The red bars indicate positively selected sites. Maximum likelihood estimates of synonymous (α) and non-synonymous rates (β) at each site are shown as bars. The base line shows the estimates under the null model (α = β)
Fig. 2
The secondary and tertiary structures of IRF9 with a focus on Val219 detected by multiple tests as a positively selected site. a The secondary structure of IRF9 and regions around the site Val219 based on the prediction of PSIPRED in human and rat orthologues. b The tertiary structure of IRF9 in human and rat orthologues based on the inference by Alphafold2. The black arrows show the site inferred to be under positive selection. The two stars (**) show the probability of the prediction of PSIPRED (> 99%)
Fig. 3
The ancestral state reconstruction and cross-species orthologous alignment. a Ancestral state reconstruction for human Val129 (red arrow) based on the Maximum Parsimony (MP) method in MEGA11. b Regional alignment comprising the human Val129 site (black arrow above the alignment)
Fig. 4
Structural similarities among homologous IRF proteins. a A simplified scheme of the IRF9 domain, showing DBD, a linker region, and IAD; b The nucleotide similarities between IRF9 and other homologous genes. The positively selected site Val129 is shown with a black arrow. The horizontal axis in b indicates CDS coordinates. The vertical axis shows the similarity between IRF9 and its homologs. c The enlarged regional structural alignment of IRF9 and IRF3 around the positively-selected site 129 of human IRF9. The 3D alignment between IRF9 and IRF3 was computed with PyMol v2.5. The 3D structures were based on Alphafold2 modeling
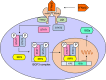
Fig. 5
Schematic molecular processes of the type I IFN signalling based on a literature review. The abbreviations and full names are IFNAR1 (Interferon Alpha and Beta Receptor Subunit 1), IFNAR2 (Interferon Alpha and Beta Receptor Subunit 2), TYK2 (Tyrosine Kinase 2), JAK (Janus kinase), STAT1 (Signal Transducer and Activator Of Transcription 1), STAT2 (Signal Transducer And Activator Of Transcription 2), ISRE (IFN stimulated response elements), and ISGs (interferon-stimulated genes)
Similar articles
- Expression regulation of zebrafish interferon regulatory factor 9 by promoter analysis.
Shi J, Zhang YB, Zhang JS, Gui JF. Shi J, et al. Dev Comp Immunol. 2013 Dec;41(4):534-43. doi: 10.1016/j.dci.2013.07.017. Epub 2013 Aug 1. Dev Comp Immunol. 2013. PMID: 23916490 - The evolution and functional characterization of miiuy croaker interferon regulatory factor 9 involved in immune response.
Yang Q, Cui J, Song W, Zhao X, Xu T. Yang Q, et al. Fish Shellfish Immunol. 2017 Jul;66:524-530. doi: 10.1016/j.fsi.2017.05.053. Epub 2017 May 22. Fish Shellfish Immunol. 2017. PMID: 28546020 - Expression and functional characterization of interferon regulatory factors (irf2, irf7 and irf9) in the blunt snout bream (Megalobrama amblycephala).
Zhan FB, Liu H, Lai RF, Jakovlić I, Wang WM. Zhan FB, et al. Dev Comp Immunol. 2017 Feb;67:239-248. doi: 10.1016/j.dci.2016.09.014. Epub 2016 Sep 25. Dev Comp Immunol. 2017. PMID: 27677680 - The emerging role of interferon regulatory factor 9 in the antiviral host response and beyond.
Suprunenko T, Hofer MJ. Suprunenko T, et al. Cytokine Growth Factor Rev. 2016 Jun;29:35-43. doi: 10.1016/j.cytogfr.2016.03.002. Epub 2016 Mar 4. Cytokine Growth Factor Rev. 2016. PMID: 26987614 Review. - A Positive Feedback Amplifier Circuit That Regulates Interferon (IFN)-Stimulated Gene Expression and Controls Type I and Type II IFN Responses.
Michalska A, Blaszczyk K, Wesoly J, Bluyssen HAR. Michalska A, et al. Front Immunol. 2018 May 28;9:1135. doi: 10.3389/fimmu.2018.01135. eCollection 2018. Front Immunol. 2018. PMID: 29892288 Free PMC article. Review.
Cited by
- Evolutionarily new genes in humans with disease phenotypes reveal functional enrichment patterns shaped by adaptive innovation and sexual selection.
Chen JH, Landback P, Arsala D, Guzzetta A, Xia S, Atlas J, Sosa D, Zhang YE, Cheng J, Shen B, Long M. Chen JH, et al. bioRxiv [Preprint]. 2024 Sep 4:2023.11.14.567139. doi: 10.1101/2023.11.14.567139. bioRxiv. 2024. PMID: 38045239 Free PMC article. Preprint.
References
- Barreiro LB, Quintana-Murci L. From evolutionary genetics to human immunology: how selection shapes host defence genes. Nat Rev Genet. 2010;11(1):17–30. - PubMed
- Zhao S, Zhang T, Liu Q, Wu H, Su B, Shi P, Chen H. Identifying lineage-specific targets of natural selection by a bayesian analysis of genomic polymorphisms and divergence from multiple species. Mol Biol Evol. 2019;36(6):1302–1315. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous