Analysis of Whole-Genome Sequences of Pathogenic Gram-Positive and Gram-Negative Isolates from the Same Hospital Environment to Investigate Common Evolutionary Trends Associated with Horizontal Gene Exchange, Mutations and DNA Methylation Patterning - PubMed (original) (raw)
Analysis of Whole-Genome Sequences of Pathogenic Gram-Positive and Gram-Negative Isolates from the Same Hospital Environment to Investigate Common Evolutionary Trends Associated with Horizontal Gene Exchange, Mutations and DNA Methylation Patterning
Ilya S Korotetskiy et al. Microorganisms. 2023.
Abstract
Hospital-acquired infections are a generally recognized problem for healthcare professionals. Clinical variants of Gram-negative and Gram-positive pathogens are characterized with enhanced antibiotic resistance and virulence due to mutations and the horizontal acquisition of respective genetic determinants. In this study, two Escherichia coli, two Klebsiella pneumoniae, three Pseudomonas aeruginosa, two Staphylococcus aureus, one Staphylococcus epidermidis and one Streptococcus pneumoniae showing broad spectra of antibiotic resistance were isolated from patients suffering from nosocomial infections in a local hospital in Almaty, Kazakhstan. The aim of the study was to compare general and species-specific pathways of the development of virulence and antibiotic resistance through opportunistic pathogens causing hospital-acquired infections. The whole-genome PacBio sequencing of the isolates allowed for the genotyping and identification of antibiotic resistance and virulence genetic determinants located in the chromosomes, plasmids and genomic islands. It was concluded that long-read sequencing is a useful tool for monitoring the epidemiological situation in hospitals. Marker antibiotic resistance mutations common for different microorganisms were identified, which were acquired due to antibiotic-selective pressure in the same clinical environment. The genotyping and identification of strain-specific DNA methylation motifs were found to be promising in estimating the risks associated with hospital infection outbreaks and monitoring the distribution and evolution of nosocomial pathogens.
Keywords: MLST; PacBio sequencing; bacterial pathogen; genotyping; hospital infection; methylomics; virulence factor.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
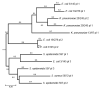
Figure 1
Genealogical tree of plasmids of the selected isolates.
Figure 2
(A) Clustering of GIs identified in different genomes of pathogenic isolates through similarities in their patterns of tetranucleotides. Each node on the scheme corresponds to one GI. Numbers in several nodes are the consecutive clockwise numbers of GIs identified in the genomes, starting from the replication origins. Host microorganisms are depicted using different colors, as explained in the legend. (B) The network of GIs is identical to that in part A, but recolored in a way that the darker grey color represents more recent inserts of GIs, i.e., the GIs with more dissimilar patterns of tetranucleotides compared to the patterns calculated for the host microorganisms. The gradient bar below the scheme shows the correspondence of the grey shading to the distance between patterns calculated with the SeqWord Sniffer program.
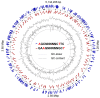
Figure 3
Atlas presentation of the circular chromosome of the strain E. coli 3/145. Locations of motifs methylated at both DNA strands are depicted with colored triangles, as explained in the legend.
Similar articles
- Whole Genome Sequencing of Extended Spectrum β-lactamase (ESBL)-producing Klebsiella pneumoniae Isolated from Hospitalized Patients in KwaZulu-Natal, South Africa.
Founou RC, Founou LL, Allam M, Ismail A, Essack SY. Founou RC, et al. Sci Rep. 2019 Apr 18;9(1):6266. doi: 10.1038/s41598-019-42672-2. Sci Rep. 2019. PMID: 31000772 Free PMC article. - Plasmids Shaped the Recent Emergence of the Major Nosocomial Pathogen Enterococcus faecium.
Arredondo-Alonso S, Top J, McNally A, Puranen S, Pesonen M, Pensar J, Marttinen P, Braat JC, Rogers MRC, van Schaik W, Kaski S, Willems RJL, Corander J, Schürch AC. Arredondo-Alonso S, et al. mBio. 2020 Feb 11;11(1):e03284-19. doi: 10.1128/mBio.03284-19. mBio. 2020. PMID: 32047136 Free PMC article. - [Analysis on distribution and drug resistance of pathogen caused community-onset bloodstream infection].
Mao S, Ge Z, Zhao H, Cao J, Xia Z. Mao S, et al. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019 Jan;31(1):67-72. doi: 10.3760/cma.j.issn.2095-4352.2019.01.014. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2019. PMID: 30707872 Chinese. - Literature review on the distribution characteristics and antimicrobial resistance of bacterial pathogens in neonatal sepsis.
Wang J, Zhang H, Yan J, Zhang T. Wang J, et al. J Matern Fetal Neonatal Med. 2022 Mar;35(5):861-870. doi: 10.1080/14767058.2020.1732342. Epub 2020 Feb 26. J Matern Fetal Neonatal Med. 2022. PMID: 32102584 Review. - The global problem of antibiotic resistance.
Gootz TD. Gootz TD. Crit Rev Immunol. 2010;30(1):79-93. doi: 10.1615/critrevimmunol.v30.i1.60. Crit Rev Immunol. 2010. PMID: 20370622 Review.
Cited by
- Special Issue "Multidrug-Resistant Bacteria in the Environment, Their Resistance and Transfer Mechanisms".
Grohmann E. Grohmann E. Microorganisms. 2023 Apr 10;11(4):981. doi: 10.3390/microorganisms11040981. Microorganisms. 2023. PMID: 37110404 Free PMC article. - Interplay of intracellular and trans-cellular DNA methylation in natural archaeal consortia.
Reva ON, La Cono V, Crisafi F, Smedile F, Mudaliyar M, Ghosal D, Giuliano L, Krupovic M, Yakimov MM. Reva ON, et al. Environ Microbiol Rep. 2024 Apr;16(2):e13258. doi: 10.1111/1758-2229.13258. Environ Microbiol Rep. 2024. PMID: 38589217 Free PMC article. - Enzyme Production and Inhibitory Potential of Pseudomonas aeruginosa: Contrasting Clinical and Environmental Isolates.
Aqel H, Sannan N, Foudah R, Al-Hunaiti A. Aqel H, et al. Antibiotics (Basel). 2023 Aug 23;12(9):1354. doi: 10.3390/antibiotics12091354. Antibiotics (Basel). 2023. PMID: 37760651 Free PMC article.
References
- Boyd S., Vasudevan A., Moore L., Brewer C., Gilchrist M., Costelloe C., Gordon A., Holmes A. Validating a prediction tool to determine the risk of nosocomial multidrug-resistant Gram-negative bacilli infection in critically ill patients: A retrospective case-control study. J. Glob. Antimicrob. Resist. 2020;22:826–831. doi: 10.1016/j.jgar.2020.07.010. - DOI - PubMed
- Ozenen G., Bal Z., Umit Z., Avcu G., Tekin D., Kurugol Z., Cilli F., Ozkinay F. Nosocomial non-fermentative Gram negative bacteria bloodstream infections in children; Risk factors and clinical outcomes of carbapenem resistance. J. Infect. Chemother. 2021;27:729–735. doi: 10.1016/j.jiac.2020.12.024. - DOI - PubMed
- Khan H., Ahmad A., Mehboob R. Nosocomial infections and their control strategies. Asian Pac. J. Trop. Biomed. 2015;5:509–514. doi: 10.1016/j.apjtb.2015.05.001. - DOI
- Jenkins D. Nosocomial infections and infection control. Medicine. 2017;45:629–633. doi: 10.1016/j.mpmed.2017.07.005. - DOI
LinkOut - more resources
Full Text Sources