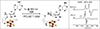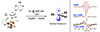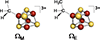Mechanism of Radical Initiation in the Radical SAM Enzyme Superfamily - PubMed (original) (raw)
Review
Mechanism of Radical Initiation in the Radical SAM Enzyme Superfamily
Brian M Hoffman et al. Annu Rev Biochem. 2023.
Abstract
Radical _S_-adenosylmethionine (SAM) enzymes use a site-differentiated [4Fe-4S] cluster and SAM to initiate radical reactions through liberation of the 5'-deoxyadenosyl (5'-dAdo•) radical. They form the largest enzyme superfamily, with more than 700,000 unique sequences currently, and their numbers continue to grow as a result of ongoing bioinformatics efforts. The range of extremely diverse, highly regio- and stereo-specific reactions known to be catalyzed by radical SAM superfamily members is remarkable. The common mechanism of radical initiation in the radical SAM superfamily is the focus of this review. Most surprising is the presence of an organometallic intermediate, Ω, exhibiting an Fe-C5'-adenosyl bond. Regioselective reductive cleavage of the SAM S-C5' bond produces 5'-dAdo• to form Ω, with the regioselectivity originating in the Jahn-Teller effect. Ω liberates the free 5'-dAdo• as the catalytically active intermediate through homolysis of the Fe-C5' bond, in analogy to Co-C5' bond homolysis in B12, which was once viewed as biology's choice of radical generator.
Keywords: 5′-deoxyadenosyl radical; S-adenosylmethionine; organometallic; radical SAM.
Figures
Figure 1.
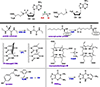
Radical SAM reactions are initiated by the reductive cleavage of SAM to generate methionine and a 5′-dAdo• radical that abstracts H• from a substrate (RH) to produce a substrate radical R• (top). The substrates and their radical transformations vary widely, with six representative reactions shown (bottom). RS enzyme names are in blue and substrates are labeled in purple. GRE-AE, glycyl radical enzyme activating enzyme; LipA, lipoyl synthase; SPL, spore photoprodut lyase; HemN, coproporphyrinogen III synthase; HydG, hydrogenase maturase; TYW1, tRNA modifying enzyme.
Figure 2.
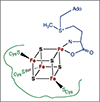
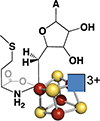
SAM coordinates to the unique iron of the [4Fe-4S] cluster via the amino and carboxylate groups. SAM is shown in blue and the protein cysteinate ligands in green.
Figure 3.
PFL-AE has a specific cation binding site near the catalytic [4Fe-4S] cluster, bridged to the cluster by the SAM carboxylate. SAM is shown in teal and the cation K+ in purple.
Figure 4.
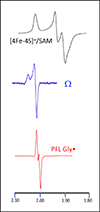
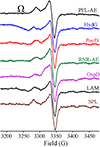
EPR spectra of the [4Fe-4S]+/SAM, Ω, and Gly• species.
Figure 5.
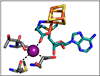
The Ω intermediate trapped by RFQ.
Figure 6.
Ω is formed in a wide array of radical SAM enzymes. Reprinted with permission from ref. 21. Copyright 2018 American Chemical Society.
Figure 7.
Coenzyme B12 (left) and the structurally analogous Ω (right). Both harbor organometallic M-C bonds to an adenosyl moiety, and both liberate a reactive 5′-dAdo• species by homolytic cleavage of the M-C bond.
Figure 8.
Photoinduced electron transfer in the PFL-AE [4Fe-4S]+/SAM complex results in reductive S-C5′ bond cleavage to generate the 5’-dAdo• radical, as revealed by EPR. Adapted with permission from ref. 40. Copyright 2019 American Chemical Society.
Figure 9.
Photoinduced electron transfer in the HydG [4Fe-4S]+/SAM complex results in reductive S-CH3 bond cleavage to generate a •CH3 radical, as revealed by EPR. Adapted with permission from ref. 42. Copyright 2019 American Chemical Society.
Figure 10.
Organometallic species methyl-omega (ΩM) and ethyl-omega (ΩE) observed upon photolysis of radical SAM enzymes with SAM and SAE.
Figure 11.
SAM ribose conformations in radical SAM enzyme active sites, either 2’-endo or 3’-endo, correlate with photoinduced reductive cleavage regioselectivity (left). Regioselectivity is introduced by active-site forces that favor one Jahn-Teller distortion, with a single elongated S-C bond that is favored for cleavage (right). Adapted with permission from ref. 44. Copyright 2021 American Chemical Society.
Figure 12.
Reaction of PFL-AE with SAM and Dha-pep gives Ω (left) followed by 5′-dAdo• (center), and then the product Ado-Dha-pep• radical (right). Adapted with permission from ref. 46. Copyright 2022 American Chemical Society.
Figure 13.
Mechanistic model for radical initiation in RS enzymes, with the three sequential intermediates observed in recent work boxed in purple
Similar articles
- Radical SAM enzymes: Nature's choice for radical reactions.
Broderick JB, Broderick WE, Hoffman BM. Broderick JB, et al. FEBS Lett. 2023 Jan;597(1):92-101. doi: 10.1002/1873-3468.14519. Epub 2022 Oct 27. FEBS Lett. 2023. PMID: 36251330 Free PMC article. Review. - Mechanism of Radical Initiation in the Radical S-Adenosyl-l-methionine Superfamily.
Broderick WE, Hoffman BM, Broderick JB. Broderick WE, et al. Acc Chem Res. 2018 Nov 20;51(11):2611-2619. doi: 10.1021/acs.accounts.8b00356. Epub 2018 Oct 15. Acc Chem Res. 2018. PMID: 30346729 Free PMC article. Review. - Active-Site Controlled, Jahn-Teller Enabled Regioselectivity in Reductive S-C Bond Cleavage of _S_-Adenosylmethionine in Radical SAM Enzymes.
Impano S, Yang H, Jodts RJ, Pagnier A, Swimley R, McDaniel EC, Shepard EM, Broderick WE, Broderick JB, Hoffman BM. Impano S, et al. J Am Chem Soc. 2021 Jan 13;143(1):335-348. doi: 10.1021/jacs.0c10925. Epub 2020 Dec 29. J Am Chem Soc. 2021. PMID: 33372786 Free PMC article. - Radical SAM Enzyme Spore Photoproduct Lyase: Properties of the Ω Organometallic Intermediate and Identification of Stable Protein Radicals Formed during Substrate-Free Turnover.
Pagnier A, Yang H, Jodts RJ, James CD, Shepard EM, Impano S, Broderick WE, Hoffman BM, Broderick JB. Pagnier A, et al. J Am Chem Soc. 2020 Oct 28;142(43):18652-18660. doi: 10.1021/jacs.0c08585. Epub 2020 Oct 15. J Am Chem Soc. 2020. PMID: 32966073 Free PMC article. - ENDOR Spectroscopy Reveals the "Free" 5'-Deoxyadenosyl Radical in a Radical SAM Enzyme Active Site Actually is Chaperoned by Close Interaction with the Methionine-Bound [4Fe-4S]2+ Cluster.
Yang H, Ho MB, Lundahl MN, Mosquera MA, Broderick WE, Broderick JB, Hoffman BM. Yang H, et al. J Am Chem Soc. 2024 Feb 14;146(6):3710-3720. doi: 10.1021/jacs.3c09428. Epub 2024 Feb 3. J Am Chem Soc. 2024. PMID: 38308759 Free PMC article.
Cited by
- Proteomic strategies to interrogate the Fe-S proteome.
Bak DW, Weerapana E. Bak DW, et al. Biochim Biophys Acta Mol Cell Res. 2024 Oct;1871(7):119791. doi: 10.1016/j.bbamcr.2024.119791. Epub 2024 Jun 25. Biochim Biophys Acta Mol Cell Res. 2024. PMID: 38925478 Review. - Benzylic Radical Stabilization Permits Ether Formation During Darobactin Biosynthesis.
Woodard AM, Peccati F, Navo CD, Jiménez-Osés G, Mitchell DA. Woodard AM, et al. bioRxiv [Preprint]. 2023 Nov 29:2023.11.29.569256. doi: 10.1101/2023.11.29.569256. bioRxiv. 2023. PMID: 38076856 Free PMC article. Preprint. - DPH1 Gene Mutations Identify a Candidate SAM Pocket in Radical Enzyme Dph1•Dph2 for Diphthamide Synthesis on EF2.
Ütkür K, Schmidt S, Mayer K, Klassen R, Brinkmann U, Schaffrath R. Ütkür K, et al. Biomolecules. 2023 Nov 16;13(11):1655. doi: 10.3390/biom13111655. Biomolecules. 2023. PMID: 38002337 Free PMC article. - Pyruvate formate-lyase activating enzyme: The catalytically active 5'-deoxyadenosyl radical caught in the act of H-atom abstraction.
Lundahl MN, Yang H, Broderick WE, Hoffman BM, Broderick JB. Lundahl MN, et al. Proc Natl Acad Sci U S A. 2023 Nov 21;120(47):e2314696120. doi: 10.1073/pnas.2314696120. Epub 2023 Nov 13. Proc Natl Acad Sci U S A. 2023. PMID: 37956301 Free PMC article. - Darobactin Substrate Engineering and Computation Show Radical Stability Governs Ether versus C-C Bond Formation.
Woodard AM, Peccati F, Navo CD, Jiménez-Osés G, Mitchell DA. Woodard AM, et al. J Am Chem Soc. 2024 May 22;146(20):14328-14340. doi: 10.1021/jacs.4c03994. Epub 2024 May 10. J Am Chem Soc. 2024. PMID: 38728535 Free PMC article.
References
- Sofia HJ, Chen G, Hetzler BG, Reyes-Spindola JF, Miller NE. 2001. Radical SAM, a novel protein superfamily linking unresolved steps in familiar biosynthetic pathways with radical mechanisms: functional characterization using new analysis and information visualization methods. Nucleic Acids Res. 29: 1097–106 - PMC - PubMed
- Cheek J, Broderick JB. 2001. Adenosylmethionine-dependent iron-sulfur enzymes: versatile clusters in a radical new role. J. Biol. Inorg. Chem. 6: 209–26 - PubMed
- Oberg N, Precord TW, Mitchell DA, Gerlt JA. 2022. RadicalSAM.org: A resource to interpret sequence-function space and discover new radical SAM chemistry. ACS Bio Med Chem Au 2: 22–35 - PMC - PubMed
- Nicolet Y 2020. Structure-function relationships of radical of radical SAM enzymes. Nat. Catal. 3: 337–2020
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous