The genomic diversity of Taiwanese Austronesian groups: Implications for the "Into- and Out-of-Taiwan" models - PubMed (original) (raw)
The genomic diversity of Taiwanese Austronesian groups: Implications for the "Into- and Out-of-Taiwan" models
Dang Liu et al. PNAS Nexus. 2023.
Abstract
The origin and dispersal of the Austronesian language family, one of the largest and most widespread in the world, have long attracted the attention of linguists, archaeologists, and geneticists. Even though there is a growing consensus that Taiwan is the source of the spread of Austronesian languages, little is known about the migration patterns of the early Austronesians who settled in and left Taiwan, i.e. the "Into-Taiwan" and "out-of-Taiwan" events. In particular, the genetic diversity and structure within Taiwan and how this relates to the into-/out-of-Taiwan events are largely unexplored, primarily because most genomic studies have largely utilized data from just two of the 16 recognized Highland Austronesian groups in Taiwan. In this study, we generated the largest genome-wide data set of Taiwanese Austronesians to date, including six Highland groups and one Lowland group from across the island and two Taiwanese Han groups. We identified fine-scale genomic structure in Taiwan, inferred the ancestry profile of the ancestors of Austronesians, and found that the southern Taiwanese Austronesians show excess genetic affinities with the Austronesians outside of Taiwan. Our findings thus shed new light on the Into- and Out-of-Taiwan dispersals.
Keywords: Austronesian; Into-Taiwan; Out-of-Taiwan; Taiwan; genome-wide data.
© The Author(s) 2023. Published by Oxford University Press on behalf of National Academy of Sciences.
Figures
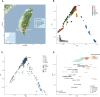
Fig. 1.
Map of the sampled Taiwanese groups and PCA of the modern/ancient individuals from South Asia, East Asia, and Oceania. A) Sampling locations of the Taiwanese populations. Map tiles by Stamen Design (CC BY 3.0), data by OpenStreetMap (ODbL). The Atayal_HO and Amis_HO are from published Human Origins data. The small map panel on the bottom-left indicates Taiwan (green) in the geographical context of East Asia. B) PCA of the modern individuals from South Asia, East Asia, and Oceania in the merged data set, colored by regions. The dashed circle indicates where the Taiwanese individuals fall. The eigenvalues from PC1 to PC10 are shown on the bottom-left. C) PCA with the ancient individuals projected onto the PCA in B) with the modern individuals in gray. Symbol shapes indicate region and colors indicate age (D). Zoom-in of C) with a focus on the modern groups from sEA and ISEA and the ancient groups from sEA and Lapita-related Oceania, colored by languages. The position of a group is the median of the positions of individuals from the group.
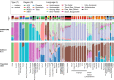
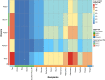
Fig. 2.
K = 9 of ADMIXTURE and K = 8 of DyStruct for selected representative groups. Both number of K are the best-fitting K for ADMIXTURE/DyStruct (Fig. S2). From the top to the bottom, there is a legend for the following three rows of keys indicating the language (L), the region (R), and the type of ancient/modern genomes (T) for the populations; below the keys, there are two rows showing the results of ADMIXTURE and DyStruct, respectively; the last row indicates the specific population label. Each vertical thin bar represents an individual, and different populations are separated with gaps. Representative groups are selected based on their enrichment of a source component, e.g. Andamanese, Indo-European, Papuan (Trans-New Guinea, East New Britain, South Bougainville), Sinto-Tibetan/nEA, Hmong-Mien, Austroasiatic, or their relevance to the into-/out-of-Taiwan events, e.g. Tai-Kadai, sEA, Taiwan, ISEA, and Oceania. Results of the full data set are presented in Fig. S3.
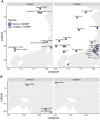
Fig. 3.
Structure within Taiwan. A) Zoom-in of the PCA plot in Fig. 1B with a focus on the Taiwanese individuals, labeled by populations and colored by groups. B) FineStructure clustering of the Sino-Tibetan, Tai-Kadai, and Austronesian groups based on haplotype painting profiles from ChromoPainter. Each vertical line represents an individual, colored according to regions (R), Taiwan groups (TG), and languages (L). Populations are labeled at the bottom. Note that the clustering does not necessarily imply phylogenetic structure. C) PCA for which eigenvalues were computed using only the THI/TOI individuals, with the Han and Lowland individuals projected. Individuals are labeled by groups and the median positions of individuals of each group are plotted with dots colored by groups. The eigenvalues from PC1 to PC10 are shown on the top-left. TW_N, TW_C, and TW_S denote northern, central, and southern Taiwanese Highland groups, respectively.
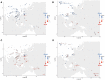
Fig. 4.
qpAdm modeling of the spatiotemporal dynamic of nEA versus sEA ancestries in East Asia and Oceania. Using outgroups shown in Fig. S8, selected groups from (A) nEA and sEA as well as (B) ISEA and Oceania (with a focus on those related to the early Out-of-Taiwan groups) are modeled with nEA (in blue, the ∼8.2 kya Boshan as proxy) and sEA (in pink, the ∼7.7 kya Liangdao as proxy) sources. The left panels show ancient groups while the right panels show modern groups. Bar plot visualization with the standard errors of the estimates is shown in Fig. S9.
Fig. 5.
Differential allelic sharing to the modern/ancient groups from East Asia and Oceania between the Atayal and Rukai/Amis. Results of the form _f_4(Atayal, X; Y, Mbuti) where X is Amis (A and B)/Rukai (C and D) and Y are modern (A and C)/ancient (B and D) groups from East Asia and Oceania. The modern/ancient groups are plotted as dots on the map, colored in proportion to Z score. Positive values (in blue) indicate more sharing with the Atayal while negative values (in red) indicate more sharing with the Amis/Rukai. Significant values (absolute Z score value ≥ 2) are further labeled with population names. For the comparisons with ancient groups, additional tests using only transversions and the French as an outgroup are shown in Fig. S12, which reduce not only the potential for false positives caused by DNA damage and/or attraction to deep outgroups but also the statistical power due to the decreased number of SNPs.
Fig. 6.
Chromosomal painting of the Malayo-Polynesian Austronesian recipients by the Formosan Austronesian donors. Results show the painting profile of recipients (in columns) by donors (in rows) colored in proportion to the average of the summed length (c
m
) of painted chromosomal regions, corresponding to the extent of haplotype sharing. The Taiwan group (TG) for the donors and the region (R) for the recipients are denoted by the colored keys. TW_N, TW_C, and TW_S indicate northern, central, and southern Taiwanese Highland groups, respectively.
Similar articles
- Early Austronesians: into and out of Taiwan.
Ko AM, Chen CY, Fu Q, Delfin F, Li M, Chiu HL, Stoneking M, Ko YC. Ko AM, et al. Am J Hum Genet. 2014 Mar 6;94(3):426-36. doi: 10.1016/j.ajhg.2014.02.003. Am J Hum Genet. 2014. PMID: 24607387 Free PMC article. - Paternal genetic affinity between Western Austronesians and Daic populations.
Li H, Wen B, Chen SJ, Su B, Pramoonjago P, Liu Y, Pan S, Qin Z, Liu W, Cheng X, Yang N, Li X, Tran D, Lu D, Hsu MT, Deka R, Marzuki S, Tan CC, Jin L. Li H, et al. BMC Evol Biol. 2008 May 15;8:146. doi: 10.1186/1471-2148-8-146. BMC Evol Biol. 2008. PMID: 18482451 Free PMC article. - Diversity and distribution of mitochondrial DNA in non-Austronesian-speaking Taiwanese individuals.
Lin M, Trejaut JA. Lin M, et al. Hum Genome Var. 2023 Jan 18;10(1):2. doi: 10.1038/s41439-022-00228-3. Hum Genome Var. 2023. PMID: 36653363 Free PMC article. - Ethnicity-based classifications and medical genetics: One Health approaches from a Western Pacific perspective.
Edinur HA, Mat-Ghani SNA, Chambers GK. Edinur HA, et al. Front Genet. 2022 Sep 6;13:970549. doi: 10.3389/fgene.2022.970549. eCollection 2022. Front Genet. 2022. PMID: 36147511 Free PMC article. Review. - Ancient migration routes of Austronesian-speaking populations in oceanic Southeast Asia and Melanesia might mimic the spread of nasopharyngeal carcinoma.
Trejaut J, Lee CL, Yen JC, Loo JH, Lin M. Trejaut J, et al. Chin J Cancer. 2011 Feb;30(2):96-105. doi: 10.5732/cjc.010.10589. Chin J Cancer. 2011. PMID: 21272441 Free PMC article. Review.
Cited by
- Multiple Human Population Movements and Cultural Dispersal Events Shaped the Landscape of Chinese Paternal Heritage.
Wang M, Huang Y, Liu K, Wang Z, Zhang M, Yuan H, Duan S, Wei L, Yao H, Sun Q, Zhong J, Tang R, Chen J, Sun Y, Li X, Su H, Yang Q, Hu L, Yun L, Yang J, Nie S, Cai Y, Yan J, Zhou K, Wang C; 10K_CPGDP Consortium; Zhu B, Liu C, He G. Wang M, et al. Mol Biol Evol. 2024 Jul 3;41(7):msae122. doi: 10.1093/molbev/msae122. Mol Biol Evol. 2024. PMID: 38885310 Free PMC article. - Investigating demic versus cultural diffusion and sex bias in the spread of Austronesian languages in Vietnam.
Thao DH, Dinh TH, Mitsunaga S, Duy D, Phuong NT, Anh NP, Anh NT, Duc BM, Hue HTT, Ha NH, Ton ND, Hübner A, Pakendorf B, Stoneking M, Inoue I, Duong NT, Hai NV. Thao DH, et al. PLoS One. 2024 Jun 17;19(6):e0304964. doi: 10.1371/journal.pone.0304964. eCollection 2024. PLoS One. 2024. PMID: 38885215 Free PMC article.
References
- Eberhard DM, Simons GF, Fennig CD. 2021. Ethnologue: languages of the world. 24th ed. Dallas (TX): SIL International.
- Blust R. 1999. Subgrouping, circularity, and extinction: some issues in Austronesian comparative linguistics. Selected Papers From the Eighth International Conference on Austronesian Linguistics; Institute of Linguistics, Academia Sinica, Taipei, pp. 31–94.
- Gray RD, Drummond AJ, Greenhill SJ. 2009. Language phylogenies reveal expansion pulses and pauses in Pacific settlement. Science 323:479–483. - PubMed
- Diamond J, Bellwood P. 2003. Farmers and their languages: the first expansions. Science 300:597–603. - PubMed
- Bellwood P. 2011. Holocene population history in the pacific region as a model for worldwide food producer dispersals. Curr Anthropol. 52:S363–S378.
LinkOut - more resources
Full Text Sources