Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans - PubMed (original) (raw)
. 2024 Mar 15;383(6688):eadj9223.
doi: 10.1126/science.adj9223. Epub 2024 Mar 15.
Sarah Moraïs 1 2 3, Sarah Winkler 1 2 3, Alvah Zorea 1 2 3, Falk S P Nagies 5, Nils Kapust 5, Eva Lamed 6, Avital Artan-Furman 6, David N Bolam 7, Madhav P Yadav 8, Edward A Bayer 1 2 6, William F Martin 5, Itzhak Mizrahi 1 2 3
Affiliations
- PMID: 38484069
- PMCID: PMC7615765
- DOI: 10.1126/science.adj9223
Cryptic diversity of cellulose-degrading gut bacteria in industrialized humans
Sarah Moraïs et al. Science. 2024.
Abstract
Humans, like all mammals, depend on the gut microbiome for digestion of cellulose, the main component of plant fiber. However, evidence for cellulose fermentation in the human gut is scarce. We have identified ruminococcal species in the gut microbiota of human populations that assemble functional multienzymatic cellulosome structures capable of degrading plant cell wall polysaccharides. One of these species, which is strongly associated with humans, likely originated in the ruminant gut and was subsequently transferred to the human gut, potentially during domestication where it underwent diversification and diet-related adaptation through the acquisition of genes from other gut microbes. Collectively, these species are abundant and widespread among ancient humans, hunter-gatherers, and rural populations but are rare in populations from industrialized societies thus indicating potential disappearance in response to the westernized lifestyle.
Conflict of interest statement
Conflicts of interest: The authors declare no conflicts of interest.
Figures
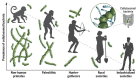
Cellulose degrading gut bacteria of hominids across evolutionary time.
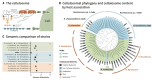
Fig. 1. Detection of a human-gut, fiber-degrading ruminococcal species.
(A) Scheme of cellulosome architecture. The CttA protein by virtue of its CBMs mediates the binding of the bacterial cell to the cellulosic substrate which can be hydrolyzed by dockerin-bearing enzymatic units that are integrated into the cell-surface cellulosome through its cohesin-containing scaffoldin assemblies. (B) Unrooted phylogenetic tree computed with the maximum likelihood method of 62 selected genomes and MAGs using the sequence of the ScaC scaffoldin illustrated in Fig. 1A as a phylotyping marker (15, 16) (table S1). The color of the clade indicates the origin of the genomic bin (light blue, human; light green, rumen). Light purple circles on the branches represent bootstrap values higher than 60%. The number and composition of cellulosomal elements is indicated as a bar for each genomic bin (number of dockerin-containing proteins with additional CAZyme elements, dark gray; number of dockerin-containing proteins with no additional CAZyme elements, medium gray; number of scaffoldins containing at least one cohesin module, light gray). Brown circles next to the MAG name indicate genomes containing a cttA gene. (C) Genomic dissimilarity computed by Mash distance within the identified ruminococcal cellulosomal species and pairwise comparisons to each other as well as to the ruminal R. flavefaciens species and the human species R. champanellensis.
Fig. 2. Ruminococcus spp. are abundant in ancient human, hunter-gatherer, and rural populations.
(A) Observed collective prevalence of the MAGs for fiber-degrading strains in various human, ape, and NHP cohorts. Pie charts represent the observed prevalences. (B) Worldwide locations of positive human and NHP samples. The locations of the samples in which the human MAGs were detected are denoted on the map as circles: dark blue, industrialized societies; light blue, rural societies and hunter-gatherers; green, paleofeces; and pink, wild NHP. (C) Distribution of fibrolytic strains in human and NHP populations. (i) Stacked bar chart of the distribution of each human cellulosomal strain (R. champanellensis, R. hominiciens, R. ruminiciens, and R. primaciens) across the sample cohorts. (ii) Heatmap of the distribution of the human cellulosomal strains among the human- and NHP-positive samples. The bar plot above the heatmap represents the number of strains detected in each sample.
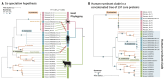
Fig. 3. Colonization by ruminococci is ongoing and dynamic in the human gut.
(A) Core protein phylogenetic tree illustrating the cospeciation hypothesis (left panel). Blue circles on the branches represent bootstrap values higher than 60%. The comparison with the phylogenetic tree of the mammalian host species is given on the right with red lines indicating proteins that do not recapitulate host phylogeny. (B) Phylogenetic tree of 197 concatenated core proteins. Blue circles on the branches represent bootstrap values higher than 77%. Blue highlighting on the right indicates a close phylogenetic distance between the human and ruminant clades. In (A) and (B) MAGs are color-coded according to host origin: green, blue, or pink indicate rumen, human, or NHP, respectively; transitional strains are denoted as “recent transfers” and the tree scales represent the number of amino acid substitutions per site. MAGs corresponding to Ruminococcus flavefaciens are indicated.
Fig. 4. Cellulosome assembly activity and cellulose adhesion.
(A) Summary of interactions between selected cellulosomal recombinant cohesin and dockerin modules derived from an R. primaciens strain (Human_SRR5558136_bin.38) compared with those of orthologous modules from the R. flavefaciens FD-1 rumen strain (79). Cohesin and dockerin modules are color-coded (red, yellow, or green) according to their predicted specificities of interaction. On both panels, light blue highlights negative interactions; darker blue, positive interactions; gray, not tested. On the left panel (R. primaciens), intensities of the interactions are denoted with − for no affinity, (OD450 lower than 0.15), + for moderate affinity (OD450 between 0.15 and 0.5), ++ for high affinity (OD450 between 0.5 and 1.0), and +++ for very high affinity (OD450 between 1.0 and 2.2), respectively. On the right panel (R. flavefaciens), intensities were not available for the Israeli-Ruimy 2017 study. (B) Overview of cellulosomal interactions in R. primaciens compared with those of R. flavefaciens as deduced from affinity-based ELISA experiments and proposed recognition residues of the dockerin components (table S6). (C) Comparative cellulolytic activity of ruminococcal GH5 orthologs of either human (R. primaciens) or rumen origin (R. flavefaciens FD-1). Enzyme samples were examined using microcrystalline cellulose (Avicel) as the substrate at 37°C. The data points represent the average of biological triplicates with standard deviation. (D) Cellulose binding assay. SDS-PAGE gels loaded with cellulose-bound (B) and -unbound (U) fractions of either R. hominiciens CttA, the CBM3a from the CipA scaffoldin of the Clostridium thermocellum cellulosome as a positive control or green fluorescent protein (GFP) as a negative control (nonbinding protein).
Fig. 5. Functional adaptation of MAGs with their host.
In (A), (C), (D), and (E), MAGs and samples are color-coded according to host origin: green, blue, or pink indicating rumen, human, or NHP, respectively. (A) Principal component analysis (PCA) of the overall predicted ORFs of the MAGs, color-coded by their hosts (see below). Clustering analysis of MAG gene content according to their hosts was performed using the PERMANOVA test with 1000 randomizations of the data and the _P_-value is indicated. (B) Rank distribution of verticality values for core proteins across the three host types versus host-specific proteins indicates that specific genes are likely to be transferred through horizontal gene transfer within a given type of host. (C) PCA of the fibrolytic system [indicating glycoside hydrolase (GH) families] of the MAGs color-coded by their hosts. Clustering analysis of MAGs GH family content according to their hosts was performed using PERMANOVA test with 1000 randomizations of the data and the p-value is indicated. (D) PCA of the expression of the fibrolytic system as examined by transcriptomic analysis of three fecal samples of the three hosts (macaque, human, and sheep rumen). (E) Center panel: heatmap of the statistically significant GH families that distinguish the strains associated with the three gut ecosystems as determined by the Kruskal-Wallis test P <0.05 after FDR correction. The left bar graph represents the verticality values for each of these orthologous groups of genes. (Right) heatmap of the statistically significant GH expression (metatranscripts in FPKM) between the three types of hosts (see material and methods section). For the GH141-Doc and GH97-Doc genes, the metatranscripts were aligned to Rumen_CADBJG01 and Rumen_CACVQO01 MAG sequences (59).
Similar articles
- The Ruminococci: key symbionts of the gut ecosystem.
La Reau AJ, Suen G. La Reau AJ, et al. J Microbiol. 2018 Mar;56(3):199-208. doi: 10.1007/s12275-018-8024-4. Epub 2018 Feb 28. J Microbiol. 2018. PMID: 29492877 Review. - Interactions between gut microbes and host promote degradation of various fiber components in Meishan pigs.
Pu G, Hou L, Zhao Q, Liu G, Wang Z, Zhou W, Niu P, Wu C, Li P, Huang R. Pu G, et al. mSystems. 2025 Feb 18;10(2):e0150024. doi: 10.1128/msystems.01500-24. Epub 2025 Jan 28. mSystems. 2025. PMID: 39873521 Free PMC article. - Ultra-deep sequencing of Hadza hunter-gatherers recovers vanishing gut microbes.
Carter MM, Olm MR, Merrill BD, Dahan D, Tripathi S, Spencer SP, Yu FB, Jain S, Neff N, Jha AR, Sonnenburg ED, Sonnenburg JL. Carter MM, et al. Cell. 2023 Jul 6;186(14):3111-3124.e13. doi: 10.1016/j.cell.2023.05.046. Epub 2023 Jun 21. Cell. 2023. PMID: 37348505 Free PMC article. - Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition.
Moraïs S, Ben David Y, Bensoussan L, Duncan SH, Koropatkin NM, Martens EC, Flint HJ, Bayer EA. Moraïs S, et al. Environ Microbiol. 2016 Feb;18(2):542-56. doi: 10.1111/1462-2920.13047. Epub 2015 Oct 14. Environ Microbiol. 2016. PMID: 26347002 - Soluble Dietary Fiber, One of the Most Important Nutrients for the Gut Microbiota.
Guan ZW, Yu EZ, Feng Q. Guan ZW, et al. Molecules. 2021 Nov 11;26(22):6802. doi: 10.3390/molecules26226802. Molecules. 2021. PMID: 34833893 Free PMC article. Review.
Cited by
- Dietary fibre optimisation in support of global health.
Ramsteijn AS, Louis P. Ramsteijn AS, et al. Microb Biotechnol. 2024 Aug;17(8):e14542. doi: 10.1111/1751-7915.14542. Microb Biotechnol. 2024. PMID: 39096198 Free PMC article. Review. - SeqCode in the golden age of prokaryotic systematics.
Jiménez DJ, Rosado AS. Jiménez DJ, et al. ISME J. 2024 Jan 8;18(1):wrae109. doi: 10.1093/ismejo/wrae109. ISME J. 2024. PMID: 38896025 Free PMC article. - Nutrient acquisition strategies by gut microbes.
Muramatsu MK, Winter SE. Muramatsu MK, et al. Cell Host Microbe. 2024 Jun 12;32(6):863-874. doi: 10.1016/j.chom.2024.05.011. Cell Host Microbe. 2024. PMID: 38870902 Free PMC article. Review. - Metatranscriptomics for Understanding the Microbiome in Food and Nutrition Science.
Butowski CF, Dixit Y, Reis MM, Mu C. Butowski CF, et al. Metabolites. 2025 Mar 10;15(3):185. doi: 10.3390/metabo15030185. Metabolites. 2025. PMID: 40137150 Free PMC article. Review. - Microbiome: A Key Regulator of Body-Brain Interactions.
O'Riordan KJ, Aburto MR, Nagpal J, Clarke G, Cryan JF. O'Riordan KJ, et al. Adv Exp Med Biol. 2025;1477:139-203. doi: 10.1007/978-3-031-89525-8_6. Adv Exp Med Biol. 2025. PMID: 40442386 Review.
References
- Freeman HJ, Spiller GA, Kim YS. A double-blind study on the effect of purified cellulose dietary fiber on 1,2-dimethylhydrazine-induced rat colonic neoplasia. Cancer Res. 1978;38:2912–2917. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical