De-novo assembly of four rail (Aves: Rallidae) genomes: A resource for comparative genomics - PubMed (original) (raw)
Editorial
. 2024 Jul 18;14(7):e11694.
doi: 10.1002/ece3.11694. eCollection 2024 Jul.
Affiliations
- PMID: 39026944
- PMCID: PMC11255403
- DOI: 10.1002/ece3.11694
Editorial
De-novo assembly of four rail (Aves: Rallidae) genomes: A resource for comparative genomics
Julien Gaspar et al. Ecol Evol. 2024.
Abstract
Rails are a phenotypically diverse family of birds that includes 130 species and displays a wide distribution around the world. Here we present annotated genome assemblies for four rails from Aotearoa New Zealand: two native volant species, pūkeko Porphyrio melanotus and mioweka Gallirallus philippensis, and two endemic flightless species takahē Porphyrio hochstetteri and weka Gallirallus australis. Using the sequence read data, heterozygosity was found to be lowest in the endemic flightless species and this probably reflects their relatively small populations. The quality checks and comparison with other rallid genomes showed that the new assemblies were of good quality. This study significantly increases the number of available rallid genomes and will enable future genomic studies on the evolution of this family.
Keywords: Gallirallus australis; Gallirallus philippensis; Porphyrio hochstetteri; Porphyrio melanotus; Rallidae; genome assemblies; heterozygosity; rails.
© 2024 The Author(s). Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
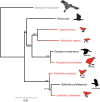
FIGURE 1
Maximum likelihood (RAxML V.8) phylogeny of three volant (black) and five flightless (red) rail lineages (Aves: Rallidae) based on 10 concatenated nuclear genes analysed with Balearica regulorum crane (Aves: Gruidae) (grey) as outgroup; bootstrap supports are indicated for each node.
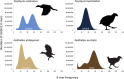
FIGURE 2
K‐mer frequency in four rails from Aotearoa New Zealand. K‐mer (nucleotide sequence of a certain length) were 57, 61, 61, 87 for Gallirallus australis, Gallirallus philippensis, Porphyrio melanotus and Porphyrio hochstetteri respectively. In each distribution, two main peaks correspond to the genomic K‐mers for the heterozygous (left) and homozygous (right) parts of the genome. The single main peak of P. hochstetteri indicates high homozygosity. Low depth peaks corresponding to erroneous K‐mer populations have been masked for clarity. Icons indicate flightless and volant species.
FIGURE 3
Average heterozygosity at 20 randomly selected genes from four newly assembled and annotated rail genomes (average total length 266,456 bp). Heterozygosity is the proportion of total nucleotide sites per individual site having two bases. Icons indicate flightless and volant species.
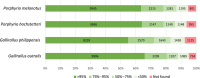
FIGURE 4
Completeness of CDSs retrieved from eight rail genomes compared to the reference crane Balearica regulorum genome that has a total of 15,035 annotated CDSs. Colours indicate the proportion of genes retrieved from a sample at various scales of completeness.
Similar articles
- Flightlessness and phylogeny amongst endemic rails (Aves:Rallidae) of the New Zealand region.
Trewick SA. Trewick SA. Philos Trans R Soc Lond B Biol Sci. 1997 Apr 29;352(1352):429-46. doi: 10.1098/rstb.1997.0031. Philos Trans R Soc Lond B Biol Sci. 1997. PMID: 9163823 Free PMC article. - Ancient mitochondrial genomes unveil the origins and evolutionary history of New Zealand's enigmatic takahē and moho.
Verry AJF, Mas-Carrió E, Gibb GC, Dutoit L, Robertson BC, Waters JM, Rawlence NJ. Verry AJF, et al. Mol Ecol. 2024 Feb;33(3):e17227. doi: 10.1111/mec.17227. Epub 2023 Nov 29. Mol Ecol. 2024. PMID: 38018770 - Testicular and spermatozoan parameters in the pukeko (Porphyrio porphyrio melanotus).
Gunn MR, Champion Z, Casey ME, Teal P, Casey PJ. Gunn MR, et al. Anim Reprod Sci. 2008 Dec;109(1-4):330-42. doi: 10.1016/j.anireprosci.2007.11.002. Epub 2007 Nov 19. Anim Reprod Sci. 2008. PMID: 18162336 - Systematics, morphology and ecology of rails (Aves: Rallidae) of the Mascarene Islands, with one new species.
Hume JP. Hume JP. Zootaxa. 2019 Jul 3;4626(1):zootaxa.4626.1.1. doi: 10.11646/zootaxa.4626.1.1. Zootaxa. 2019. PMID: 31712544 - Evolution of bird genomes-a transposon's-eye view.
Kapusta A, Suh A. Kapusta A, et al. Ann N Y Acad Sci. 2017 Feb;1389(1):164-185. doi: 10.1111/nyas.13295. Epub 2016 Dec 20. Ann N Y Acad Sci. 2017. PMID: 27997700 Review.
References
- Baker, A. J. , Daugherty, C. H. , Colbourne, R. , & McLennan, J. L. (1995). Flightless brown kiwis of New Zealand possess extremely subdivided population structure and cryptic species like small mammals. Proceedings of the National Academy of Sciences of the United States of America, 92, 8254–8258. - PMC - PubMed
- Boast, A. P. , Chapman, B. , Herrera, M. B. , Worthy, T. H. , Scofield, R. P. , Tennyson, A. J. D. , Houde, P. , Bunce, M. , Cooper, A. , & Mitchell, K. J. (2019). Mitochondrial genomes from New Zealand's extinct adzebills (Aves: Aptornithidae: Aptornis) support a sister‐taxon relationship with the Afro‐Madagascan Sarothruridae. Diversity, 11, 24.
Publication types
LinkOut - more resources
Full Text Sources