Genome-wide characterization of 54 urinary metabolites reveals molecular impact of kidney function - PubMed (original) (raw)
Meta-Analysis
Genome-wide characterization of 54 urinary metabolites reveals molecular impact of kidney function
Erkka Valo et al. Nat Commun. 2025.
Abstract
Dissecting the genetic mechanisms underlying urinary metabolite concentrations can provide molecular insights into kidney function and open possibilities for causal assessment of urinary metabolites with risk factors and disease outcomes. Proton nuclear magnetic resonance metabolomics provides a high-throughput means for urinary metabolite profiling, as widely applied for blood biomarker studies. Here we report a genome-wide association study meta-analysed for 3 European cohorts comprising 8,011 individuals, covering both people with type 1 diabetes and general population settings. We identify 54 associations (p < 9.3 × 10-10) for 19 of 54 studied metabolite concentrations. Out of these, 33 were not reported previously for relevant urinary or blood metabolite traits. Subsequent two-sample Mendelian randomization analysis suggests that estimated glomerular filtration rate causally affects 13 urinary metabolite concentrations whereas urinary ethanolamine, an initial precursor for phosphatidylcholine and phosphatidylethanolamine, was associated with higher eGFR lending support for a potential protective role. Our study provides a catalogue of genetic associations for 53 metabolites, enabling further investigation on how urinary metabolites are linked to human health.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: P-H G has served on advisory boards for AbbVie, Astellas, AstraZeneca, Bayer, Boehringer Ingelheim, Cebix, Eli Lilly, Janssen, Medscape, MSD, Mundipharma, Nestlé, Novartis, Novo Nordisk, Sanofi, and has received lecture honoraria from Astellas, AstraZeneca, Bayer, Berlin Chemie, Boehringer Ingelheim, Eli Lilly, Elo Water, Genzyme, Medscape, Menarini, MSD, Mundipharma, Novartis, Novo Nordisk PeerVoice Sanofi, and Sciarc. P-H G has also received investigator-initiated grants from Eli Lilly and Roche. S M received lecture honoraria from Encore Medical Education. All other authors declare no competing interests.
Figures
Fig. 1. An overview of the genome-wide characterization of the urinary metabolites.
We performed genome-wide association study (GWAS) meta-analyses in a total of 8,011 individuals from three cohorts to investigate genetic variants associated with 54 urinary metabolites measured by NMR. Conditional and joint analysis (COJO) was applied to identify independent secondary signals within the associated genetic loci. The identified associations were characterized by expression quantitative trait loci (eQTL) analysis and association look-ups from GWAS catalogue and kidney GWAS data. We performed two-directional Mendelian Randomization to study the causal associations between the urine metabolites and health outcomes. Pathway analyses were performed to obtain wider understanding of the biological pathways affecting each metabolite. FinnDiane: Finnish Diabetic Nephropathy Study. GS: Generation Scotland. SNV: single nucleotide variant. Meta-GWAS two-sided p-values calculated with METAL applying inverse variance weighted method and genomic control correction for the individual study level results. Conditional and joint analysis two-sided p-values calculated with GCTA-COJO applying conditional and joint analysis of independently associated variants. eGFR: estimated glomerular filtration rate. UACR: urinary albumin creatinine ratio. DKD: diabetic kidney disease. T2D: type 2 diabetes. Created in BioRender. Valo, E. (2024) BioRender.com/a61e932.
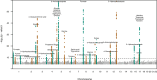
Fig. 2. Manhattan plot of signals with p < 5.0×10−5 for the metabolites.
Signals from different metabolites are clumped together if they are within 50 kb from another signal. Pruned genome-wide significant signals with p < 5 × 10−8/54 = 9.3 × 10−10 and variants 1 Mb around them are highlighted with two alternating colours. Note: y-axis clipped at 60. P-values calculated with METAL applying inverse variance weighted method and genomic control correction for the individual study level results. All p-values are two-sided.
Fig. 3. Regional association with tyrosine for lead variants rs11133665 and rs7704882 on chromosome 5.
Upper panel shows LocusZoom plot centred around the previously known rs11133665 variant, and the previously unreported signal at rs7704882 independently associated with tyrosine. P-values were calculated with METAL applying inverse variance weighted method and genomic control correction for the individual study level results. The middle panel shows kidney eQTL associations for SLC6A18 and SLC6A19 overlaid on top of the tyrosine association signals, highlighting lead variants rs11133665 (eQTL for SLC6A19 in kidney_)_ and rs7704882 (eQTL for SLC6A18 in the kidney_)_. The lower panel contains the protein-coding genes in the region with exons highlighted. The r 2 values with rs11133665 are calculated based on 1000 Genomes phase 3 European population. Variants with no r 2 information are not shown. P-values from two-sided tests.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous