Bacterially expressed F1-20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission - PubMed (original) (raw)
Bacterially expressed F1-20/AP-3 assembles clathrin into cages with a narrow size distribution: implications for the regulation of quantal size during neurotransmission
W Ye et al. J Neurosci Res. 1995.
Abstract
F1-20/AP-3 is a synapse-specific phosphoprotein. In this study we characterize the ability of bacterially expressed F1-20/AP-3 to bind and assemble clathrin cages. We find that both of two bacterially expressed alternatively spliced isoforms of F1-20/AP-3 can bind and assemble clathrin as efficiently as preparations of F1-20/AP-3 from bovine brain. This establishes that the clathrin assembly activity found in F1-20/AP-3 preparations from brain extracts is indeed encoded by the cloned gene for F1-20/AP-3. It also demonstrates that post-translation modification is not required for activation of the clathrin binding or assembly function of F1-20/AP-3. Ultrastructural analyses of the clathrin cages assembled by bacterially expressed F1-20/AP-3 reveals a strikingly narrow size distribution. This may be important for the regulation of quantal size during neurotransmission. We also express the 33 kD NH2-terminus of F1-20/AP-3 in E. coli, and measure its ability to bind to clathrin triskelia, to bind to clathrin cages, and to assemble clathrin triskelia into clathrin cages. It has been suggested that the 33 kD NH2-terminus of F1-20/AP-3 constitutes a clathrin binding domain. We find that the bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 binds to clathrin triskelia, fails to bind to preassembled clathrin cages, and is not sufficient for clathrin assembly. The finding that the 33 kD NH2-terminus of F1-20/AP-3 binds to clathrin triskelia but fails to assemble clathrin triskelia into clathrin cages is consistent with the published proteolysis studies. The finding that the 33 kD NH2-terminus of F1-20/AP-3 fails to bind to clathrin cages is novel and potentially important. It is clear from these experiments that the 33 kD NH2-terminus of F1-20/AP-3 is sufficient to carry out some aspects of clathrin binding; however it appears that defining the regions of the protein involved in clathrin binding and assembly may be more complex than originally anticipated.
Figures
Fig. 1
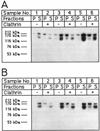
Bacterially expressed GST-F1-20/AP-3 (AS15−) and GST-F1-20/AP-3 (AS15+) binds to clathrin cages. Either 80 µg/ml (samples 1 and 2), 160 µg/ml (samples 3 and 4), or 240 µg/ml (samples 5 and 6) of either GST-F1-20/AP-3 (AS15−) (A) or F1-20/AP-3 (AS15 +) (B) were incubated in the absence (−) or presence (+) of 400 µg/ml clathrin cages for 45 min at 4°C in tartrate buffer. Following a low-speed spin to remove non-specific aggregates, the clathrin cages were pelleted by ultracentrifugation at 100,000g. The pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE, followed by western blot analysis with the F1-20 Mab. The uppermost bands are the intact fusion proteins. The lower bands are proteolytic fragments. [Note, we found that F1-20/AP-3 (AS15 +) is more susceptible to aggregation than F1-20/AP-3 (AS15−); hence even though equal amounts of protein were used in the experiments shown in A and B, more aggregates were removed in the low-speed spin in the experiment which utilized F1-20/AP-3 (AS15+) (panel B) than in the experiment which utilized F1-20/AP-3 (AS15−) (A).]
Fig. 2
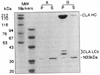
The bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 does not bind to clathrin cages; 240 µg/ml of the bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 was incubated in the absence (A) or presence (B) of 800 µg/ml clathrin cages for 45 min at 4°C in isolation buffer. Following a low-speed spin to remove non-specific aggregates, the clathrin cages were pelleted by ultracentrifugation at 100,000g. The pellet (P) and supernatant (S) fractions were analyzed by SDS-PAGE, followed by Coomassie Blue staining. The positions of clathrin heavy chain (CLA HC), clathrin light chains (CLA LCs), and the 33 kD NH2-terminus (N33kDa) are indicated. Pellets were loaded at triple concentrations. Sizes of the molecular weight markers (Pharmacia) are indicated. When this experiment was repeated with an additional two independent clones expressing the 33 kD NH2-terminus, the results were identical (data not shown).
Fig. 3
The bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 binds specifically to clathrin triskelia; 15 µg of the bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 was incubated with 0.5 ml clathrin-Sepharose in 0.5 ml isolation buffer at 4°C for 2 hr (A), and binding was monitored by batch analysis, as described in Methods. Fraction 1 is the flow-through; fractions 2,3,4 are washes with isolation buffer; and fractions 5,6,7 are eluates with 0.5 M Tris (pH 7.0). All samples were analyzed by SDS-PAGE, followed by silver staining. Negative controls were carried out by incubating 15 µg bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 with 0.5 ml underivatized Sepharose (B), and by incubating 15 µg E. coli GST protein with 0.5 ml clathrin-Sepharose (C).
Fig. 4
Bacterially expressed GST-F1-20/AP-3 (AS15−) and GST-F1-20/AP-3 (AS15+) assemble clathrin triskelia into cages, while the bacterially expressed 33 kD NH2-terminus of F1-20/AP-3 is not sufficient for clathrin assembly; 2 mg/ml clathrin triskelia were dialyzed overnight at 4°C against Isolation buffer alone (A), or with the addition of either 2 mg/ml GST (B), 2 mg/ml purified bovine brain F1-20/AP-3 (C), 2 mg/ml GST-F1-20/AP-3 (AS15−) (D), 2 mg/ml GST-F1-20/AP-3 (AS15+) (E), or 2 mg/ml 33 kD NH2-terminus of F1-20/AP-3 (F). The three test proteins at 2 mg/ml were also dialyzed overnight at 4°C against isolation buffer without clathrin triskelia: GST-F1-20/AP-3 (AS 15−) (G), GST-F1-20/AP-3 (AS15+) (H). Following a low-speed spin to remove non-specific aggregates, newly assembled clathrin cages were pelleted by ultracentrifugation at 100,000g. The pellet and supernatant fractions were analyzed by SDS-PAGE, followed by silver staining. The distribution of the clathrin light chains between the pellet and supernatant fractions were used to evaluate assembly, since the clathrin heavy chain co-migrates with F1-20/AP-3.
Fig. 5
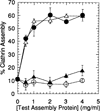
Quantitative factor-dependent assembly of clathrin triskelia; 2 mg/ml clathrin triskelia were dialyzed overnight at 4°C against isolation buffer alone, or with the addition of the indicated concentrations of either GST-F1-20/AP-3 (AS15−) (open triangle), GST-F1-20/AP-3 (AS15+) (solid circle), the 33 kD NH2-terminus of F1-20/AP-3 (solid triangle), or the E. coli GST protein (open circle). Following a low-speed spin to remove non-specific aggregates, newly assembled clathrin cages were pelleted by ultracentrifugation at 100,000g. The pellet and supernatant fractions were analyzed by SDS-PAGE, followed by silver staining. The distribution of the clathrin light chains between the pellet and supernatant fractions were quantitated using a Millipore BioImage System with 3cx scanner. Multiple scans at varied loadings were performed to ensure linearity. Each data point on the plot is the average assembly determined from three independent assembly assays. The error bars indicate one standard deviation from this average.
Fig. 6
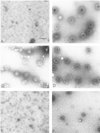
Ultrastructural analysis of assembled clathrin cages. The morphology of the products of the assembly assays depicted in Figure 3 were evaluated by negative staining electron microscopy. They were compared with the products of a factor-independent assembly assay in which 2 mM free Ca++ was added to initiate clathrin assembly (B). Products are shown from the factor-dependent assembly assays containing either no added protein (A), GST-F1-20/AP-3 (AS15−) (C), GST-F1-20/AP-3 (AS15+) (D), GST (E), or the 33 kD NH2-terminus of F1-20/AP-3 (F). Scale bar, 130 nm.
Fig. 7
Size distribution of the assembled clathrin cages. Diameters of 393 cages assembled in the factor-independent clathrin assembly assay in which 2 mM free Ca++ was added to initiate clathrin assembly were measured (top). Diameters of 306 cages assembled in the factor-dependent clathrin assembly assay with GST-F1-20/AP-3 (AS15−) were measured (bottom).
Similar articles
- Clathrin binding and assembly activities of expressed domains of the synapse-specific clathrin assembly protein AP-3.
Ye W, Lafer EM. Ye W, et al. J Biol Chem. 1995 May 5;270(18):10933-9. doi: 10.1074/jbc.270.18.10933. J Biol Chem. 1995. PMID: 7738035 Free PMC article. - The synapse-specific phosphoprotein F1-20 is identical to the clathrin assembly protein AP-3.
Zhou S, Tannery NH, Yang J, Puszkin S, Lafer EM. Zhou S, et al. J Biol Chem. 1993 Jun 15;268(17):12655-62. J Biol Chem. 1993. PMID: 7685348 - Recognition sites for clathrin-associated proteins AP-2 and AP-3 on clathrin triskelia.
Murphy JE, Keen JH. Murphy JE, et al. J Biol Chem. 1992 May 25;267(15):10850-5. J Biol Chem. 1992. PMID: 1587861 - Endocytosis: an assembly protein for clathrin cages.
McMahon HT. McMahon HT. Curr Biol. 1999 May 6;9(9):R332-5. doi: 10.1016/s0960-9822(99)80206-1. Curr Biol. 1999. PMID: 10330371 Review. - The AP-3 complex: a coat of many colours.
Odorizzi G, Cowles CR, Emr SD. Odorizzi G, et al. Trends Cell Biol. 1998 Jul;8(7):282-8. doi: 10.1016/s0962-8924(98)01295-1. Trends Cell Biol. 1998. PMID: 9714600 Review.
Cited by
- A role for an Hsp70 nucleotide exchange factor in the regulation of synaptic vesicle endocytosis.
Morgan JR, Jiang J, Oliphint PA, Jin S, Gimenez LE, Busch DJ, Foldes AE, Zhuo Y, Sousa R, Lafer EM. Morgan JR, et al. J Neurosci. 2013 May 1;33(18):8009-21. doi: 10.1523/JNEUROSCI.4505-12.2013. J Neurosci. 2013. PMID: 23637191 Free PMC article. - A Novel Sequence in AP180 and CALM Promotes Efficient Clathrin Binding and Assembly.
Moshkanbaryans L, Xue J, Wark JR, Robinson PJ, Graham ME. Moshkanbaryans L, et al. PLoS One. 2016 Aug 30;11(8):e0162050. doi: 10.1371/journal.pone.0162050. eCollection 2016. PLoS One. 2016. PMID: 27574975 Free PMC article. - Modulation of HIV-like particle assembly in vitro by inositol phosphates.
Campbell S, Fisher RJ, Towler EM, Fox S, Issaq HJ, Wolfe T, Phillips LR, Rein A. Campbell S, et al. Proc Natl Acad Sci U S A. 2001 Sep 11;98(19):10875-9. doi: 10.1073/pnas.191224698. Epub 2001 Aug 28. Proc Natl Acad Sci U S A. 2001. PMID: 11526217 Free PMC article. - Of condensates and coats - reciprocal regulation of clathrin assembly and the growth of protein networks.
Malady BT, Papagiannoula A, Kamatar A, Sarkar S, Lebrun GT, Wang L, Hayden CC, Lafer EM, Owen DJ, Milles S, Stachowiak JC. Malady BT, et al. bioRxiv [Preprint]. 2025 May 14:2025.05.13.653742. doi: 10.1101/2025.05.13.653742. bioRxiv. 2025. PMID: 40463115 Free PMC article. Preprint. - The monomeric clathrin assembly protein, AP180, regulates contractile vacuole size in Dictyostelium discoideum.
Stavrou I, O'Halloran TJ. Stavrou I, et al. Mol Biol Cell. 2006 Dec;17(12):5381-9. doi: 10.1091/mbc.e06-06-0531. Epub 2006 Oct 18. Mol Biol Cell. 2006. PMID: 17050736 Free PMC article.
References
- Bennett MK, Scheller RH. A molecular description of synaptic vesicle membrane trafficking. Annu Rev Biochem. 1994;63:63–100. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources