Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior - PubMed (original) (raw)
Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior
M H Vitaterna et al. Science. 1994.
Abstract
In a search for genes that regulate circadian rhythms in mammals, the progeny of mice treated with N-ethyl-N-nitrosourea (ENU) were screened for circadian clock mutations. A semidominant mutation, Clock, that lengthens circadian period and abolishes persistence of rhythmicity was identified. Clock segregated as a single gene that mapped to the midportion of mouse chromosome 5, a region syntenic to human chromosome 4. The power of ENU mutagenesis combined with the ability to clone murine genes by map position provides a generally applicable approach to study complex behavior in mammals.
Figures
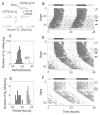
Fig. 1. ENU mutagenesis screen
A behavioral assay was used to test for the presence of circadian rhythm mutations in firstgeneration offspring of ENU-treated males. (A) The mutagenesis procedure. Young male B6 mice were injected intraperitoneally with ENU at 150 mg/kg. On recovery of fertility, they were bred with B6 females to produce G1 offspring heterozygous for any induced mutations. Dominant or semidominant mutations could then be detected in G1 offspring. (B) Representative activity record of a wild-type G1 female from the screen. The record is double-plotted so that 48 hours are shown for each horizontal trace, and each day’s record is presented both to the right and beneath that of the preceding day. Wheel revolutions are recorded as pen deflections so that the dark regions represent times of activity. The animal was kept in LD14:10 for the first 20 days (illustrated by the bar above the record), then transferred to constant darkness (DD). Light pulses of 5 min were given on the days indicated by arrows. On transfer to DD, this animal exhibited a period of 23.7 hours. (C) Distribution of period among 304 G1 offspring tested for circadian phenotype. One animal, #25, exhibited a period that deviated from the mean by more than an hour. (D) Activity record of G1-25, the founder animal. This animal exhibited a period of 24.8 hours after 30 days in DD. (E) Distribution of period among 23 B6 N2 offspring from the founder animal. Thirteen offspring exhibited periods within the normal range, whereas 10 had periods longer than normal, comparable with that of the founder animal. (F) Representative activity record of a heterozygous male B6 N2 offspring. This animal had a period of 24.8 hours in DD.
Fig. 2. Circadian activity records of F2 generation mice
The activity LD records of six F2 offspring [B6 F DD A to D; (BALB × B6)F2, E to H] are shown. All animals were kept on LD12:12 for the first 7 to 10 days illustrated, then transferred to DD on the day indicated by a line in the right margin. (A) Activity record of a wild-type B6 F2 mouse. This animal had a period of 23.6 hours in DD. (B) Activity record of a heterozygous Clock/+ B6 F2 mouse. This animal had a period of 24.8 hours in DO. (C) Activity record of a homozygous Clock/Clock B6 F2 mouse. This individual had a period of 27.1 hours for the first 10 days in DD. There is a loss of circadian rhythmicity thereafter and only an ultradian rhythm is apparent. (D) The same activity record as illustrated in (C), plotted on a 27-hour time base. The approximately 27-hour periodicity during the first part of exposure to DD appears as vertically aligned activity bouts. (E) Activity record of a wild-type (BALB × B6)F2 mouse. This individual had a steady-state period of 23.1 hours in DD. A 6-hour light pulse was given on the day indicated by an arrow, resulting in a 1-hour phase advance in the activity rhythm. (F) Activity record of a heterozygous Clock/+ (BALB × B6)F2 mouse. This individual had a steady-state period of 24.7 hours in DD. A 6-hour light pulse was given on the day indicated by an arrow, resulting in a 3.3-hour phase advance in the activity rhythm. (G) Activity record of a homozygous Clock/Clock (BALB × B6)F2 mouse. This animal had a long period during the initial interval in DD, which damped out after about five cycles leaving an ultradian pattern. After a 6-hour light pulse indicated by the arrow, a period of 28.2 hours was observed, which then persisted for at least 15 cycles. (H) The same activity record as illustrated in (G) plotted on a 28.5-hour time base to show the transient restoration of a long periodicity after the light pulse by vertically aligning activity bouts.
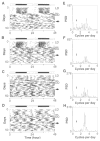
Fig. 3. Analysis of periodicity of homozygous Clock mutants
The activity records of four homozygous Clock/Clock (BALB × B6)F2 individuals are shown (A to D). All animals shown were kept under LD12:12 for the first 12 days shown, then transferred to DD as indicated by the line in the right margin. A 6-hour light pulse was given on the day indicated by an arrow. Among the animals whose records are shown, no circadian rhythmicity persisted for more than about 10 cycles after either the initial transfer to DD or the light pulse. The corresponding Fourier analysis for each activity record is shown to the right (E to H). Fourier analyses encompass a 10-day interval corresponding to the 10 days preceding the light pulse (18). PSD, power spectral density. The frequency corresponding to one cycle per day, or a 24-hour period, is indicated by an arrow. Peaks corresponding to approximately 6- to 9-hour ultradian periods are seen for each animal.
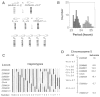
Fig. 4. Mapping of the Clock mutation
A panel of 100 [(BALB × B6)F1 × B6]N2 mice was used to map Clock by genetic linkage and haplotype analysis. (A) The mating system used to produce the backcross panel. A schematic illustration of a pair of chromosomes is shown for each animal: circles represent centromeres, squares represent alleles at loci along the length of the chromosome, open symbols represent BALB alleles, and filled symbols represent B6 alleles. Clock heterozygous (+/m) B6 males were bred with wild-type (+/+) BALB females. The offspring were behaviorally tested and the Clock heterozygous F1 individuals were backcrossed to wild-type B6 to produce the N2 generation. One chromosome of each pair of autosomes in the N2 offspring is of B6 provenance (from the B6 parent), whereas the other (from the F1 parent) may be of either parental type or recombinant. Because the Clock mutation descends from a B6 animal, N2 offspring that are Clock/+ should carry B6 alleles at loci that flank the Clock locus, while +/+ offspring should carry BALB alleles at these loci. (B) Distribution of the period phenotype among the 100 animals. The steady-state period clearly distinguished animals between the two genotypes: 58 individuals were wild type and 42 individuals were heterozygous. See Table 1, cross 7, for analysis. (C) The assortment of haplotypes for chromosome 5 observed among the 100 N2 generation mice. Thirty-six animals had nonrecombinant haplotypes, 48 had singly recombinant haplotypes, and 16 exhibited doubly recombinant haplotypes. (D) Map of chromosome 5 showing the position of Clock relative to SSLP markers. Distances between loci are shown to the left in centimorgans ± standard error (cM ± SE), calculated with the program Map Manager (31). The lod scores to the right of each locus represent the logarithm of the odds for the pairwise test of linkage of the locus to Clock.
Similar articles
- Mutagenesis and behavioral screening for altered circadian activity identifies the mouse mutant, Wheels.
Pickard GE, Sollars PJ, Rinchik EM, Nolan PM, Bucan M. Pickard GE, et al. Brain Res. 1995 Dec 24;705(1-2):255-66. doi: 10.1016/0006-8993(95)01171-4. Brain Res. 1995. PMID: 8821757 - The mouse Clock mutation behaves as an antimorph and maps within the W19H deletion, distal of Kit.
King DP, Vitaterna MH, Chang AM, Dove WF, Pinto LH, Turek FW, Takahashi JS. King DP, et al. Genetics. 1997 Jul;146(3):1049-60. doi: 10.1093/genetics/146.3.1049. Genetics. 1997. PMID: 9215907 Free PMC article. - Two kinds of ENU-induced scant hair mice and mapping of the mutant genes.
Wu BJ, Shao YX, Mao HH, Tang D, Liu J, Xue ZF, Li HD. Wu BJ, et al. J Dermatol Sci. 2004 Dec;36(3):149-56. doi: 10.1016/j.jdermsci.2004.08.010. J Dermatol Sci. 2004. PMID: 15541636 - New insights into behaviour using mouse ENU mutagenesis.
Oliver PL, Davies KE. Oliver PL, et al. Hum Mol Genet. 2012 Oct 15;21(R1):R72-81. doi: 10.1093/hmg/dds318. Epub 2012 Aug 13. Hum Mol Genet. 2012. PMID: 22892373 Free PMC article. Review. - Finding the genes that direct mammalian development : ENU mutagenesis in the mouse.
Anderson KV. Anderson KV. Trends Genet. 2000 Mar;16(3):99-102. doi: 10.1016/s0168-9525(99)01921-6. Trends Genet. 2000. PMID: 10689347 Review.
Cited by
- The role of clock in ethanol-related behaviors.
Ozburn AR, Falcon E, Mukherjee S, Gillman A, Arey R, Spencer S, McClung CA. Ozburn AR, et al. Neuropsychopharmacology. 2013 Nov;38(12):2393-400. doi: 10.1038/npp.2013.138. Epub 2013 May 31. Neuropsychopharmacology. 2013. PMID: 23722243 Free PMC article. - From Chronodisruption to Sarcopenia: The Therapeutic Potential of Melatonin.
Fernández-Martínez J, Ramírez-Casas Y, Yang Y, Aranda-Martínez P, Martínez-Ruiz L, Escames G, Acuña-Castroviejo D. Fernández-Martínez J, et al. Biomolecules. 2023 Dec 12;13(12):1779. doi: 10.3390/biom13121779. Biomolecules. 2023. PMID: 38136651 Free PMC article. Review. - An Integrated View of Potassium Homeostasis.
Gumz ML, Rabinowitz L, Wingo CS. Gumz ML, et al. N Engl J Med. 2015 Jul 2;373(1):60-72. doi: 10.1056/NEJMra1313341. N Engl J Med. 2015. PMID: 26132942 Free PMC article. Review. No abstract available. - Review: Circadian clocks and rhythms in the vascular tree.
Han Q, Bagi Z, Rudic RD. Han Q, et al. Curr Opin Pharmacol. 2021 Aug;59:52-60. doi: 10.1016/j.coph.2021.04.010. Epub 2021 Jun 7. Curr Opin Pharmacol. 2021. PMID: 34111736 Free PMC article. Review. - Cocaine self-administration behaviors in ClockΔ19 mice.
Ozburn AR, Larson EB, Self DW, McClung CA. Ozburn AR, et al. Psychopharmacology (Berl). 2012 Sep;223(2):169-77. doi: 10.1007/s00213-012-2704-2. Epub 2012 Apr 26. Psychopharmacology (Berl). 2012. PMID: 22535308 Free PMC article.
References
- Klein DC, Moore RY, Reppert SM, editors. Suprachiasmatic Nucleus: The Mind’s Clock. Oxford University Press; New York: 1991.
- Takahashi JS. Curr Opin Neurobiol. 1991;1:556. - PubMed
- Takahashi JS, Murakami N, Nikaido SS, Pratt BL, Robertson LM. Recent Prog Horm Res. 1989;45:279. - PubMed
- Turek FW. 1994;49:43. ibid. - PubMed
- Michel S, Geusz ME, Zaritsky JJ, Block GD. Science. 1993;259:239. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- T32 DC000015/DC/NIDCD NIH HHS/United States
- R01-DK40493/DK/NIDDK NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- P30-CA07175/CA/NCI NIH HHS/United States
- T32 NS071040/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases