A bipolar kinesin - PubMed (original) (raw)
A bipolar kinesin
A S Kashina et al. Nature. 1996.
Abstract
Chromosome segregation during mitosis depends on the action of the mitotic spindle, a self-organizing, bipolar protein machine which uses microtubules (MTs) and their associated motors. Members of the BimC subfamily of kinesin-related MT-motor proteins are believed to be essential for the formation and functioning of a normal bipolar spindle. Here we report that KRP130, a homotetrameric BimC-related kinesin purified from Drosophila melanogaster embryos, has an unusual ultrastructure. It consists of four kinesin-related polypeptides assembled into a bipolar aggregate with motor domains at opposite ends, analogous to a miniature myosin filament. Such a bipolar 'minifilament' could crosslink spindle MTs and slide them relative to one another. We do not know of any other MT motors that have a bipolar structure.
Figures
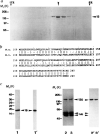
FIG. 1
Purification of KRP130 and corresponding motor-domain antibodies for electron microscopy, a, SDS-polyacrylamide gel of fractions from sucrose density gradient centrifugation of KRP130 (ref. 13). Arrowhead indicates peak fraction. Percentage sucrose (w/v) is indicated for two fractions, b, Best-fit comparison of the sea urchin (S.u.) Eg5-related motor-domain subfragment used for raising Eg5 motor-domain antibodies, and the corresponding region of Xenopus laevis (X.I.) Eg5 (identity 69%, similarity 83%). c, Characterization of the Eg5 motor-domain antibody. SDS-polyacrylamide gels (1–3) and corresponding anti-Eg5 motor-domain immunoblots (1′–3′) of a purified fusion protein of M_r 19K, (His)6-SU Eg5 motor, comprising the sea-urchin Eg5-related motor-domain fragment fused to a polyhistidine tag (1,1′), and KRP130 pelleted with MTs in the presence of the nonhydrolysable ATP analogue, 5′adenylyl-β,γ_-imidodiphosphate (AMP–PNP) (2,2′) or ATP (3,3′) in low salt. Numbers on the left refer to Mr of standards. Arrowheads indicate recombinant motor-domain fragment (19), tubulin (tub) and KRP130 (130). METHODS KRP130 was purified from Drosophila melanogaster embryos and rotary-shadowed directly or following co-pelleting with MTs in AMP–PNP or ATP (KRP130 binding to MTs is ATP-sensitive at high but not low ionic strength; Fig. 1_c and ref. 13). Antibody specific for KRP130 motor domains (Figs 1_c and 4_a_) was raised against a recombinant motor-domain fragment of a sea-urchin egg protein, KRP170, that is a close relative of frog Eg5. Expression of a KRP170 motor-domain fragment complementary DNA in pRSETB vector yielded a (His)6-SU Eg5 motor fusion protein of Mr 19K, which was purified by Ni-nitrilotriacetic acid metal-chelate affinity chromatography followed by SDS-PAGE electroelution (Fig. 1_c_, lane 1) and used to raise rabbit polyclonal antibody. Specific antibody was affinity purified from serum using the 19K fusion protein coupled to agarose, analysed by immunoblotting (Fig. lc) and used for electron microscopy (Fig. 4_a_).
FIG. 2
Electron micrographs of rotary-shadowed KPR130 molecules. These images, together with previous studies, are consistent with the hypothesis that a KRP130 holoenzyme comprises four 130K subunits assembled into a structure 96 nm long, with a total Mr of roughly 500K, Stokes radius of 16.2 nm, and S-value of 7.6 S. For comparison, kinesin comprises two heavy chains of 110K–130K and two light chains of 55K–80K assembled into a structure 75–80 nm long with a total Mr of 350K–400K, Stokes radius of 9nm, and S-value of 9.0–9.6S. The dimensions of both rotary-shadowed KRP130 and kinesin appear slightly larger than their true dimensions owing to the coating of platinum, which in our experiments is routinely estimated to be approximately 2.5 nm thick. METHODS. KRP130 alone or KRP130–MT complexes were mixed with an equal volume of 80% glycerol containing 0.6–1.0 M ammonium acetate and sprayed onto freshly cleaved mica plates. The plates were then vacuum dried, rotary shadowed with platinum at a 6° angle using a Balzers BAF 400T freeze-fracture device, and processed as described previously. The replicas were visualized on a Philips 410LS electron microscope at 80 kV. No difference in the structure of KRP130 was observed with or without the MT binding step.
FIG. 3
KRP130 molecules are bipolar structures capable of crosslinking microtubules, a, Drosophila KRP130 is probably a homologue of Xenopus Eg5, a member of the BimC subfamily, consisting of 1,060 amino acid residues arranged in a tripartite motor–rod–tail manner, according to the map (top). The model (bottom) shows how four KRP130 polypeptides could be arranged to produce bipolar homotetramers. Two parallel KRP130 dimers assemble in an antiparallel fashion to form bipolar tetramers 96 nm in length with two 10-nm motor-domain ‘heads’ protruding on each end. The ‘tails’ of the KRP130 subunits would be juxtaposed to the ‘heads’; close packing of two heads and two tails would produce a structure visible in rotary-shadowed specimens as a single, globular domain approximately 20 nm × 20 nm on each end of the 60-nm rod. This is consistent with the dimensions of the rotary-shadowed KRP130 molecules, b, Negatively stained images of complexes of KRP130 with MTs. Sometimes the ‘dumbbell-shaped’ KRP130 molecules bind by opposite ends to adjacent MTs (right), producing MT–MT crossbridges, whereas other molecules bind to MTs with one end only, producing projections from the MT wall, c, Individual fine, aperiodic cross-bridges are observed in rotary-shadowed KRP130–MT pellets (arrows). The reduced length of the crossbridges (53 nm) compared to the ‘rods’ of individual molecules (60 nm) may be due to occlusion of part of the ‘rod’ domain by MTs. Crossbridges were not observed in preparations of MTs alone (top row). METHODS. Rotary shadowing was performed as described (Fig. 2 legend). The aperiodic crossbridges are difficult to shadow because they are obscured by MTs during platinum spraying. For negative staining, KRP130–MT complexes were resuspended in buffer, loaded onto formvar-coated grids, then stained with 2% aqueous uranyl acetate for 3 min.
FIG. 4
Antibody decoration of bipolar KRP130 tetramers and models of KRP130 function, a, Rotary-shadowed KRP130 decorated with anti-Eg5 motor-domain antibody (Fig. 1_c_). About one-quarter of the molecules on the grid were undecorated (top row). Decorated molecules show the characteristic ‘dumbbell’ shape but the heads are enlarged. Because of the large size of the heads and low angle of shadowing, the contrast of the ‘rod’ of decorated molecules (middle four rows) is hard to visualize, but was clearer when rotary shadowing was prolonged to produce larger platinum grain (bottom row). b, Models for KRP130 function. In model 1, KRP130 crosslinks two antiparallel MTs emanating from opposite spindle poles with MT plus ends distal to the poles. The motor domains at opposite ends of KRP130 molecules move towards the plus ends of the crosslinked MTs, causing the MTs themselves to slide apart, minus ends leading (arrows), resulting in spindle-pole separation during the formation, maintenance or elongation of a bipolar mitotic spindle. In model 2, KRP130 could crosslink parallel MTs emanating from one or both poles and drive movements necessary for spindle formation and maintenance. For example, by moving towards the plus ends of crosslinked MTs (arrowheads) it may ‘zip’ together MTs into kinetochore fibres or, as proposed for Xenopus Eg5 (ref. 5), it could exert pushing forces (straight arrow) that drive MT flux towards the poles and take part in organizing spindle poles. We favour model one, which is more consistent with the mechanism of action of the bipolar actin-based motor, myosin II. METHODS. Purified KRP130 from the sucrose gradients was mixed with MTs and anti-Eg5 motor-domain antibody (Fig. 1) in amounts approximately equimolar to the amount of KRP130. The MT–KRP130–antibody complexes were pelleted through a glycerol cushion and processed for rotary shadowing as described (Fig. 2 legend).
Similar articles
- An essential bipolar mitotic motor.
Kashina AS, Scholey JM, Leszyk JD, Saxton WM. Kashina AS, et al. Nature. 1996 Nov 21;384(6606):225. doi: 10.1038/384225a0. Nature. 1996. PMID: 8918872 Free PMC article. No abstract available. - The bipolar kinesin, KLP61F, cross-links microtubules within interpolar microtubule bundles of Drosophila embryonic mitotic spindles.
Sharp DJ, McDonald KL, Brown HM, Matthies HJ, Walczak C, Vale RD, Mitchison TJ, Scholey JM. Sharp DJ, et al. J Cell Biol. 1999 Jan 11;144(1):125-38. doi: 10.1083/jcb.144.1.125. J Cell Biol. 1999. PMID: 9885249 Free PMC article. - A "slow" homotetrameric kinesin-related motor protein purified from Drosophila embryos.
Cole DG, Saxton WM, Sheehan KB, Scholey JM. Cole DG, et al. J Biol Chem. 1994 Sep 16;269(37):22913-6. J Biol Chem. 1994. PMID: 8083185 Free PMC article. - The bimC family of kinesins: essential bipolar mitotic motors driving centrosome separation.
Kashina AS, Rogers GC, Scholey JM. Kashina AS, et al. Biochim Biophys Acta. 1997 Jul 24;1357(3):257-71. doi: 10.1016/s0167-4889(97)00037-2. Biochim Biophys Acta. 1997. PMID: 9268050 Review. No abstract available. - Mechanisms by Which Kinesin-5 Motors Perform Their Multiple Intracellular Functions.
Pandey H, Popov M, Goldstein-Levitin A, Gheber L. Pandey H, et al. Int J Mol Sci. 2021 Jun 15;22(12):6420. doi: 10.3390/ijms22126420. Int J Mol Sci. 2021. PMID: 34203964 Free PMC article. Review.
Cited by
- Mechanisms Underlying Rare Inherited Pediatric Retinal Vascular Diseases: FEVR, Norrie Disease, Persistent Fetal Vascular Syndrome.
Le V, Abdelmessih G, Dailey WA, Pinnock C, Jobczyk V, Rashingkar R, Drenser KA, Mitton KP. Le V, et al. Cells. 2023 Nov 5;12(21):2579. doi: 10.3390/cells12212579. Cells. 2023. PMID: 37947657 Free PMC article. Review. - Regulatory mechanisms that control mitotic kinesins.
Yount AL, Zong H, Walczak CE. Yount AL, et al. Exp Cell Res. 2015 May 15;334(1):70-7. doi: 10.1016/j.yexcr.2014.12.015. Epub 2015 Jan 6. Exp Cell Res. 2015. PMID: 25576382 Free PMC article. Review. - Genome-wide RNAi screen for synthetic lethal interactions with the C. elegans kinesin-5 homolog BMK-1.
Maia AF, Tanenbaum ME, Galli M, Lelieveld D, Egan DA, Gassmann R, Sunkel CE, van den Heuvel S, Medema RH. Maia AF, et al. Sci Data. 2015 May 12;2:150020. doi: 10.1038/sdata.2015.20. eCollection 2015. Sci Data. 2015. PMID: 25984351 Free PMC article. - Bipolarization and poleward flux correlate during Xenopus extract spindle assembly.
Mitchison TJ, Maddox P, Groen A, Cameron L, Perlman Z, Ohi R, Desai A, Salmon ED, Kapoor TM. Mitchison TJ, et al. Mol Biol Cell. 2004 Dec;15(12):5603-15. doi: 10.1091/mbc.e04-05-0440. Epub 2004 Sep 22. Mol Biol Cell. 2004. PMID: 15385629 Free PMC article. - Modulation of the kinesin ATPase cycle by neck linker docking and microtubule binding.
Zhao YC, Kull FJ, Cochran JC. Zhao YC, et al. J Biol Chem. 2010 Aug 13;285(33):25213-20. doi: 10.1074/jbc.M110.123067. Epub 2010 Jun 17. J Biol Chem. 2010. PMID: 20558732 Free PMC article.
References
- Mitchison TJ. Phil. Trans. R. Soc. Lond. 1992;B336:99–106. - PubMed
- Vernos I, Karsenti E. Trends Cell Biol. 1995;5:297–301. - PubMed
- Enos AP, Morris NR. Cell. 1990;60:1019–1027. - PubMed
- Sawin KE, LeGuellec K, Philippe M, Mitchison TJ. Nature. 1992;359:540–543. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases