The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species - PubMed (original) (raw)
The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species
J A Eisen. J Mol Evol. 1995 Dec.
Abstract
The evolution of the RecA protein was analyzed using molecular phylogenetic techniques. Phylogenetic trees of all currently available complete RecA proteins were inferred using multiple maximum parsimony and distance matrix methods. Comparison and analysis of the trees reveal that the inferred relationships among these proteins are highly robust. The RecA trees show consistent subdivisions corresponding to many of the major bacterial groups found in trees of other molecules including the alpha, beta, gamma, delta, epsilon proteobacteria, cyanobacteria, high-GC gram-positives, and the Deinococcus-Thermus group. However, there are interesting differences between the RecA trees and these other trees. For example, in all the RecA trees the proteins from gram-positive species are not monophyletic. In addition, the RecAs of the cyanobacteria consistently group with those of the high-GC gram-positives. To evaluate possible causes and implications of these and other differences phylogenetic trees were generated for small-subunit rRNA sequences from the same (or closely related) species as represented in the RecA analysis. The trees of the two molecules using these equivalent species-sets are highly congruent and have similar resolving power for close, medium, and deep branches in the history of bacteria. The implications of the particular similarities and differences between the trees are discussed. Some of the features that make RecA useful for molecular systematics and for studies of protein evolution are also discussed.
Figures
Figure 1
Alignment of complete RecA sequences. The alignment was generated using the clustalw multiple sequence alignment program. Dashes (-) represent alignment gaps. Three insertions that are present in only one sequence each (Myb.t, Myb.l, and Tg.m) and the first 80 aa of the A. thaliana protein are left out for space reasons and are indicated by a ••. Conservation of alignment positions as determined by the clustalw program is indicated by * (identical aa in all) and. (similar aa in all). The alignment positions used in phylogenetic analysis are indicated by the sequence mask (1=used, 0=not used). Sequence abbreviations are described in Table 1.
Figure 1
Alignment of complete RecA sequences. The alignment was generated using the clustalw multiple sequence alignment program. Dashes (-) represent alignment gaps. Three insertions that are present in only one sequence each (Myb.t, Myb.l, and Tg.m) and the first 80 aa of the A. thaliana protein are left out for space reasons and are indicated by a ••. Conservation of alignment positions as determined by the clustalw program is indicated by * (identical aa in all) and. (similar aa in all). The alignment positions used in phylogenetic analysis are indicated by the sequence mask (1=used, 0=not used). Sequence abbreviations are described in Table 1.
Figure 2
Comparison of consensus trees for RecA and SS-rRNA. Strict-rule consensus trees representing the phylogenetic patterns found in all trees generated by multiple methods for each molecule are shown. The RecA consensus (A) was generated from the PAUP, protpars, Fitch-Margoliash, De Soete and neighbor-joining trees (see Methods). The SS-rRNA consensus (B) was generated from the dnapars, Fitch-Margoliash, De Soete and neighbor-joining trees. Comparable species are aligned in the middle and species are ordered to minimize branch crossing (note two crossed branches in SS-rRNA tree). Consensus clades are shaded for each molecule.
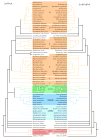
Figure 3
Fitch-Margoliash trees for RecA (A) and SS-rRNA (B). Trees were generated from the multiple sequence alignments by the method of Fitch and Margoliash. Regions of ambiguous alignment and indels were excluded from the analysis (see Methods). For the RecA tree, distances were calculated using the protdist program of PHYLIP with a PAM-matrix based distance correction. For the SS-rRNA tree, distances were calculated using the dnadist program of PHYLIP and the Kimura-2-parameter distance correction. Consensus clades representing groups found in all phylogenetic methods are highlighted. Branch lengths and scale bars correspond to estimated evolutionary distance. Bootstrap values when over 40 are indicated.
Figure 3
Fitch-Margoliash trees for RecA (A) and SS-rRNA (B). Trees were generated from the multiple sequence alignments by the method of Fitch and Margoliash. Regions of ambiguous alignment and indels were excluded from the analysis (see Methods). For the RecA tree, distances were calculated using the protdist program of PHYLIP with a PAM-matrix based distance correction. For the SS-rRNA tree, distances were calculated using the dnadist program of PHYLIP and the Kimura-2-parameter distance correction. Consensus clades representing groups found in all phylogenetic methods are highlighted. Branch lengths and scale bars correspond to estimated evolutionary distance. Bootstrap values when over 40 are indicated.
Similar articles
- Genetic diversity and phylogenetic relationships of bacteria belonging to the Ochrobactrum-Brucella group by recA and 16S rRNA gene-based comparative sequence analysis.
Scholz HC, Al Dahouk S, Tomaso H, Neubauer H, Witte A, Schloter M, Kämpfer P, Falsen E, Pfeffer M, Engel M. Scholz HC, et al. Syst Appl Microbiol. 2008 Mar;31(1):1-16. doi: 10.1016/j.syapm.2007.10.004. Epub 2008 Jan 28. Syst Appl Microbiol. 2008. PMID: 18222618 - Evolution of the recA gene and the molecular phylogeny of bacteria.
Lloyd AT, Sharp PM. Lloyd AT, et al. J Mol Evol. 1993 Oct;37(4):399-407. doi: 10.1007/BF00178869. J Mol Evol. 1993. PMID: 8308907 - Analysis of recombinase A (recA/RecA) in the actinobacterial family Streptosporangiaceae and identification of molecular signatures.
Meyers PR. Meyers PR. Syst Appl Microbiol. 2015 Dec;38(8):567-77. doi: 10.1016/j.syapm.2015.10.001. Epub 2015 Oct 22. Syst Appl Microbiol. 2015. PMID: 26507967 - Bacterial phylogeny based on 16S and 23S rRNA sequence analysis.
Ludwig W, Schleifer KH. Ludwig W, et al. FEMS Microbiol Rev. 1994 Oct;15(2-3):155-73. doi: 10.1111/j.1574-6976.1994.tb00132.x. FEMS Microbiol Rev. 1994. PMID: 7524576 Review.
Cited by
- Phylogenetic analysis of the rplA genes encoding ribosomal protein L1.
Klucar L, Nováková R, Homérová D, Sevcíková B, Turna J, Kormanec J. Klucar L, et al. Folia Microbiol (Praha). 2001;46(2):99-106. doi: 10.1007/BF02873585. Folia Microbiol (Praha). 2001. PMID: 11501409 - Burkholderia and Paraburkholderia are Predominant Soybean Rhizobial Genera in Venezuelan Soils in Different Climatic and Topographical Regions.
Ramírez MDA, España M, Aguirre C, Kojima K, Ohkama-Ohtsu N, Sekimoto H, Yokoyama T. Ramírez MDA, et al. Microbes Environ. 2019 Mar 30;34(1):43-58. doi: 10.1264/jsme2.ME18076. Epub 2019 Feb 15. Microbes Environ. 2019. PMID: 30773514 Free PMC article. - Annotation-free delineation of prokaryotic homology groups.
Yin Y, Ogilvie HA, Nakhleh L. Yin Y, et al. PLoS Comput Biol. 2022 Jun 8;18(6):e1010216. doi: 10.1371/journal.pcbi.1010216. eCollection 2022 Jun. PLoS Comput Biol. 2022. PMID: 35675326 Free PMC article. - Sequencing of heat shock protein 70 (DnaK) homologs from Deinococcus proteolyticus and Thermomicrobium roseum and their integration in a protein-based phylogeny of prokaryotes.
Gupta RS, Bustard K, Falah M, Singh D. Gupta RS, et al. J Bacteriol. 1997 Jan;179(2):345-57. doi: 10.1128/jb.179.2.345-357.1997. J Bacteriol. 1997. PMID: 8990285 Free PMC article. - Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation.
Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Eren AM, et al. PLoS One. 2011;6(10):e26732. doi: 10.1371/journal.pone.0026732. Epub 2011 Oct 25. PLoS One. 2011. PMID: 22046340 Free PMC article.
References
- Aigle B, Schneider D, Decaris B. Genbank entry Z30324. 1994.
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous