Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression - PubMed (original) (raw)
Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression
C Connelly et al. Cell. 1996.
Abstract
The budding yeast SKP1 gene, identified as a dosage suppressor of a known kinetochore protein mutant, encodes an intrinsic 22.3 kDa subunit of CBF3, a multiprotein complex that binds centromere DNA in vitro. Temperature-sensitive mutations in SKP1 define two distinct phenotypic classes. skp1-4 mutants arrest predominantly as large budded cells with a G2 DNA content and short mitotic spindle, consistent with a role in kinetochore function. skp1-3 mutants, however, arrest predominantly as multiply budded cells with a G1 DNA content, suggesting an additional role during the G1/S phase. Identification of Skp1p homologs from C. elegans, A. thaliana, and H. sapiens indicates that SKP1 is evolutionarily highly conserved. Skp1p therefore represents an intrinsic kinetochore protein conserved throughout eukaryotic evolution and may be directly involved in linking kinetochore function with the cell cycle-regulatory machinery.
Figures
Figure 1
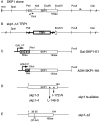
SKP1 Genomic DNA, Mutant Alleles, and Epitope Tag Fusion Proteins (A) Restriction map of the cloned SKP1 gene and flanking regions. The vector multiple cloning site (mcs) represents the original left end of the library clone where a genomic Sau3A site was ligated to a BamHI site in the vector. (B) The _skp1-Δ_1 allele was constructed by replacement of the 0.73 kb PstI–Eco57I segment with a 1.8 kb TRP1 gene containing fragment (shown in bold). (C) Schematic structures of the E1 and HA amino-terminal epitope-tagged Skp1p fusion constructs. The E1–Skp1p fusion is under transcriptional control of the GAL1 promotor; the HA–Skp1p fusion is under the transcriptional control of the ADH2 promotor. (D) Schematic of the skp1 temperature-sensitive alleles used in phenotypic studies. (E) Schematic of _skp1-Δ_2. The 28 amino acid in-frame deletion corresponds to amino acid residues 37–64.
Figure 2
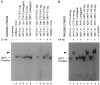
Mobility Shift Analysis of Skp1p in CBF3–CEN DNA Complexes DNA–protein complexes formed with 32P-labeled CDEIII probe and whole-cell extracts were analyzed on a nondenaturing polyacrylamide gel (Doheny et al. 1993). Antibodies were added to preformed complexes, and samples were incubated for 20 min at room temperature before gel analysis. Unbound probe was run off the bottom of the gel. (A) Lanes 1, 2, and 11 are controls. Lanes 3–6, extracts from cells containing E1-tagged or untagged Ctf13p or Skp1p proteins demonstrating an intrinsic mobility shift due to the 60 amino acid epitope fusion in each case. Lanes 7–10, extracts from cells containing E1-tagged Ctf13p or Skp1p subsequently incubated with or without antibodies directed against the E1 epitope as indicated. Lanes 7 and 9 are controls for lanes 8 and 10. (B) Lanes 1–6, extracts containing either HA-tagged or untagged Ctf13p or Skp1p subsequently incubated with or without an antibody directed against the HA epitopeas indicated. Lanes 7 and 8 are from an independent transformant (repeat of lanes 5 and 6). Lane 9 is a control.
Figure 3
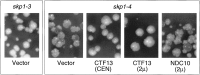
Chromosome Segregation Phenotypes of skp1-3 and skp1-4 Mutants Yeast strains carrying integrated skp1 temperature-sensitive alleles were tested for chromosome missegregation phenotypes using the colony color sectoring assay (Koshland and Hieter 1987). Loss of the nonessential marker chromosome gives a red sector in a white colony. skp1-3 mutants exhibit a wild-type phenotype (very rarely sectored colonies) when transformed with vector only. skp1-4 mutants exhibit dramatically increased rates of chromosome loss (highly sectored colonies) when transformed with vector only. Introduction of the CTF13 gene on a low copy (CEN) vector or high copy (2μ) vector suppresses the chromosome missegregation phenotype of the skp1-4 mutant. Introduction of the NDC10 gene on a 2μ vector has no effect.
Figure 4
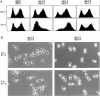
Phenotypic Analysis of skp1 Temperature-Sensitive Mutants (A) Shown are G1 arrest (at nonpermissive temperature) of skp1-3 cells and G2/M accumulation (at permissive temperature) and G2/M arrest (at nonpermissive temperature) of skp1-4 cells, as analyzed by flow cytometry (Gerring et al. 1990). The number of cells is depicted on the vertical axis, with fluorescent intensity of emitted light (proportional to DNA content) on the horizontal axis. Logarithmically growing cultures at 25°C were split and incubated at either 25°C or 37°C for 3 hr prior to analysis. (B) Terminal arrest morphologies of skp1-3 or skp1-4 cells after 3 or 6 hr at 37°C. Cells were stained with 4′,6-diamidino-2-phenylindole and photographed for fluorescence staining and by phase contrast. The fields shown were chosen to emphasize specific phenotypes and are not necessarily representative of overall frequencies.
Figure 5
Quantitation of Cell and Nuclear Morphology SKP1/SKP1 wild-type, skp1-3/skp1-3 and skp1-4/skp1-4 homozygous mutants, and skp1-3/skp1-4 heteroallelic diploids were grown to logarithmic phase at 25°C and shifted to 37°C for 3 and 6 hr, and nuclear and bud morphology were scored. The criteria used for each morphologic class scored are shown schematically above the columns. The numbers shown represent percentages of the total cells scored (far right column). Asterisk, in this population of cells, 67% had the nucleus at the neck and 33% had the nucleus spanning the neck (n = 203).
Figure 6
Multiple Protein Sequence Alignments of Skp1p Homologs The seven Skp1p homologs, from the six species indicated and the chlorella virus genome (PBCV-1), are aligned and ordered by decreasing similarity to the human protein. The consensus is derived from positions that have a 5 out of 7 identical match or better. Hs, H. sapiens; Cp, C. porcellus; Dd, D. discoideum; At, A. thaliana; Sc, S. cerevisiae; CV, PBCV-1 chlorella virus; Ce, C. elegans. Asterisks mark the positions of temperature-sensitive mutations.
Similar articles
- SGT1 encodes an essential component of the yeast kinetochore assembly pathway and a novel subunit of the SCF ubiquitin ligase complex.
Kitagawa K, Skowyra D, Elledge SJ, Harper JW, Hieter P. Kitagawa K, et al. Mol Cell. 1999 Jul;4(1):21-33. doi: 10.1016/s1097-2765(00)80184-7. Mol Cell. 1999. PMID: 10445024 - Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore.
Stemmann O, Neidig A, Köcher T, Wilm M, Lechner J. Stemmann O, et al. Proc Natl Acad Sci U S A. 2002 Jun 25;99(13):8585-90. doi: 10.1073/pnas.082223899. Proc Natl Acad Sci U S A. 2002. PMID: 12084919 Free PMC article. - Identification of essential components of the S. cerevisiae kinetochore.
Doheny KF, Sorger PK, Hyman AA, Tugendreich S, Spencer F, Hieter P. Doheny KF, et al. Cell. 1993 May 21;73(4):761-74. doi: 10.1016/0092-8674(93)90255-o. Cell. 1993. PMID: 8500169 Free PMC article. - The Saccharomyces cerevisiae kinetochore.
Lechner J, Ortiz J. Lechner J, et al. FEBS Lett. 1996 Jun 24;389(1):70-4. doi: 10.1016/0014-5793(96)00563-7. FEBS Lett. 1996. PMID: 8682209 Review. - The role of centromere-binding factor 3 (CBF3) in spindle stability, cytokinesis, and kinetochore attachment.
Bouck D, Bloom K. Bouck D, et al. Biochem Cell Biol. 2005 Dec;83(6):696-702. doi: 10.1139/o05-161. Biochem Cell Biol. 2005. PMID: 16333320 Review.
Cited by
- Molecular interaction map of the mammalian cell cycle control and DNA repair systems.
Kohn KW. Kohn KW. Mol Biol Cell. 1999 Aug;10(8):2703-34. doi: 10.1091/mbc.10.8.2703. Mol Biol Cell. 1999. PMID: 10436023 Free PMC article. Review. - The Arabidopsis gene tardy asynchronous meiosis is required for the normal pace and synchrony of cell division during male meiosis.
Magnard JL, Yang M, Chen YC, Leary M, McCormick S. Magnard JL, et al. Plant Physiol. 2001 Nov;127(3):1157-66. Plant Physiol. 2001. PMID: 11706195 Free PMC article. - Centromere position in budding yeast: evidence for anaphase A.
Guacci V, Hogan E, Koshland D. Guacci V, et al. Mol Biol Cell. 1997 Jun;8(6):957-72. doi: 10.1091/mbc.8.6.957. Mol Biol Cell. 1997. PMID: 9201708 Free PMC article. - A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae.
Guacci V, Koshland D, Strunnikov A. Guacci V, et al. Cell. 1997 Oct 3;91(1):47-57. doi: 10.1016/s0092-8674(01)80008-8. Cell. 1997. PMID: 9335334 Free PMC article. - Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast.
Patton EE, Willems AR, Sa D, Kuras L, Thomas D, Craig KL, Tyers M. Patton EE, et al. Genes Dev. 1998 Mar 1;12(5):692-705. doi: 10.1101/gad.12.5.692. Genes Dev. 1998. PMID: 9499404 Free PMC article.
References
- Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper J.W, Elledge S.J. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell, this issue. 1996 - PubMed
- Boeke J, Truehart J, Natsoulis G, Fink G. 5-Fluoro-orotic acid as a selective agent in yeast molecular genetics. Meth. Enzymol. 1987;154:164–175. - PubMed
- Boguski M, Tolstochev C, Bassett D. Gene discovery in dbEST. Science. 1994;265:1993–1994. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases