Identification of the homologous beige and Chediak-Higashi syndrome genes - PubMed (original) (raw)
. 1996 Jul 18;382(6588):262-5.
doi: 10.1038/382262a0.
Q A Nguyen, V T Tchernev, J A Ashley, J C Detter, S M Blaydes, S J Brandt, D Chotai, C Hodgman, R C Solari, M Lovett, S F Kingsmore
Affiliations
- PMID: 8717042
- PMCID: PMC2893578
- DOI: 10.1038/382262a0
Identification of the homologous beige and Chediak-Higashi syndrome genes
M D Barbosa et al. Nature. 1996.
Erratum in
- Nature 1997 Jan 2;385(6611):97
Abstract
Vesicular transport to and from the lysosome and late endosome is defective in patients with Chediak-Higashi syndrome (CHS) and in mutant beige (bg) mice. CHS and bg cells have giant, perinuclear vesicles with characteristics of late endosomes and lysosomes that arise from dysregulated homotypic fusion. CHS and bg lysosomes also exhibit compartmental missorting of proteins, such as elastase, glucuronidase and cathepsin G. Lyst, a candidate gene for bg, was identified by direct complementary DNA selection from a yeast artificial chromosome (YAC) clone containing a 650-kilobase segment of the bg-critical region on mouse chromosome 13. Lyst is disrupted by a 5-kilobase deletion in bg mice, and Lyst messenger RNA is markedly reduced in bg homozygotes. The homologous human gene, LYST, is highly conserved with mouse Lyst, and contains a frame-shift mutation at nucleotides 117-118 of the coding domain in a CHS patient. Thus bg mice and human CHS patients have homologous disorders associated with Lyst mutations. Lyst encodes a protein with a carboxy-terminal prenylation motif and multiple potential phosphorylation sites. Lyst protein is predicted to form extended helical domains, and has a region of sequence similar to stathmin, a coiled-coil phosphoprotein thought to act as a relay integrating cellular signal response coupling.
Figures
FIG. 1
Genetic and physical map of the bg non-recombinant interval on mouse chromosome 13 showing the location of Lyst. Mouse chromosome 13 is shown by a horizontal line with the centromere on the left. The bg critical region is delineated by chromosome crossovers (denoted with an X) in animals 134 and 137 of an intersub-specific mouse backcross [C57BL/6J – bgJ × (C57BL/6J – bgJ × CAST/EiJ)F1]. Microsatellite markers D13MM172 and D13Mit239 flank bg proximally; D13Mitl62 and D13Mit305 lie distal to bg (indicated by turquoise circles). YAC and P1 clones identified by PCR screening, with oligo-nucleotides corresponding to Nid or D13Sfkl3 are shown above the chromosome. Novel sequence-tagged sites (STS, indicated by dark blue circles), generated by inverse repetitive element PCR or direct cDNA selection, were used to order clones within the contiguous array. Novel mouse chromosome 13 STSs are numbered 1–18, corresponding to D13Sfkl to D13Sfkl8, respectively. Lyst was isolated from YAC 195A8, a 650-kb clone, by using direct cDNA selection. The physical location of _Lyst_-associated STSs on YAC and P1 clones are shown in red (MGD accession number MGD-PMEX-13).
FIG. 2
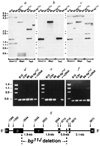
Intragenic deletion of Lyst in bg11J DNA. a–c, Southern blot identification of an intragenic Lyst deletion in bg11J DNA. A Southern blot was sequentially hybridized with 3 Lyst probes: a, the probe (nucleotides 1,262–3,433 of Lyst cDNA) extends upstream from the bg11J deletion; b, the probe (nucleotides 2,835–3,433 of Lyst cDNA) is completely deleted; c, the probe (nucleotides 3,594–4,237 of Lyst cDNA) extends downstream from the bg11J deletion. Restriction endonucleases are indicated at the bottom of each panel, and molecular size standards (in kb) are shown to the left. Similar results were obtained with 3 additional restriction endonucleases (data not shown). The bg11J mutation was discovered in 1992 at The Jackson Laboratory in a C57BL/6J-jb mouse at generation N4 after transfer from B6C3Fe-a/a. The mutation jb had, in turn, been discovered 14 generations earlier in B6C3Fe-a/a-hyh mice at generation N3 after transfer form C57BL/10J. The hyh mutation arose in C57BL710J mice, and was maintained in that strain until transfer at F15. Thus the possible contributors of genetic information to bg11J include C57BL/6J, C3HeB/FeJ and C57BL/10J. Southern blots were prepared from genomic DNA of all potential progenitor mouse strains, but only C57B/V10J, C57BL/6J and C57BI/6J-bg11J are shown, d–f, PCR analysis of the bg11J deletion. C57BL/10J, C3HeB/FeJ, C57BL/6J and C57BL/6J-bg11J genomic DNA and Lyst cDNA were used as templates in the PCR reactions. Amplicons illustrated correspond to: d, Lyst cDNA nucleotides 1,337–1,837, which represent exon β and are upstream from the deletion; e, nucleotides 2,670–3,210, which represent exon γ, which is deleted in bg11J DNA; and f, nucleotides 4,913–5,433, which represents an exon downstream from the deletion. No amplicon was observed in control PCR reactions performed without template. More than 30 other STSs that had been localized within the bg non-recombinant interval PCR amplified normally from bg11J DNA. g, Genomic structure of Lyst in the vicinity of the bg11J deletion. Lyst exons (α, β, γ, δ, ε and φ) are depicted by black boxes, and intervening introns by a solid line. Nucleotides of the mouse Lyst cDNA that correspond to exonic boundaries are indicated above the boxes. The 3′ end of exon β, and all of exons γ and δ, are deleted in bg11J DNA. The region of Lyst protein that is deleted in bg11J contains a pair of helices with N-terminal phosphorylation sites. Genomic structure and intronic sequences were ascertained by sequence analysis of nested PCR products, performed with exonic primers and P1 clone DNA as template. Boundaries of the bg11J deletion were determined by PCR of genomic DNA.
FIG. 3
Amino-acid sequence and predicted peptide structure of mouse and human Lyst cDNAs. a, Predicted amino-acid sequence of the mouse Lyst cDNA. The region (residues 463–536) with sequence similarity to stathmin is underlined, as is a C-terminal prenylation motif, b, Predicted secondary structure– and motifs of mouse Lyst protein. The solid line denotes the protein sequence. Above the line are the stathmin-like region and areas rich in acid (A; Asp, Glu), basic (B; His, Arg, Lys), and turn (T; Pro, Gly, Ser, Thr, Asp) residues, the latter, especially at regions 600–625 and 875–895, are too long to be a simple turn between secondary structural elements, and are either very flexible or very rigid (in a manner reminiscent of collagen, although they do not fit the collagen consensus motifs). Helices predicted with >80% confidence are shown as solid lines below the protein sequence, and asterisks identify helices with N-terminal phosphorylation motifs. A tyrosine phosphorylation motif (residue 1,214) is designated Y. c, Nuceiotide and predicted amino-acid sequence of the 5′ end of the human LYST coding domain showing a frame-shift mutation in a patient with CHS. Line A illustrates the predicted normal human amino-acid sequence; line B shows the normal human nucleotide sequence; line C shows the nucleotide sequence from a patient with CHS; line D shows the predicted amino-acid sequence from the CHS patient. The CHS nucleotide sequence contains an insertional mutation at nucleotide 117–118 that results in a frame shift and premature termination (asterisk).
FIG. 4
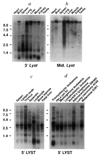
Northern blot analysis of mouse and human Lyst. a, b, Northern blots of 2 µg poly(A)+ RNA from various mouse tissues (Clontech) hybridized with probes that correspond to a, nucleotides 4,423–4,631, and b, nucleotides 1,430–2,457 (exon β) of mouse Lyst cDNA. c, d, Northern blot of 2µg poly(A)+ RNA from c, various human lymphoid tissues, or d, human cancer cell lines, hybridized with a probe that corresponds to nucleotides 357–800 of human LYST cDNA. Molecular size standards (in kb) are shown to the left. Hybridization of mouse mRNA with probes from mouse Lyst exons α and γ gave identical results to those shown with exon β (b), whereas probes from exons δ, ε, and ϕ gave results identical to those shown in a.
Similar articles
- Grey, a novel mutation in the murine Lyst gene, causes the beige phenotype by skipping of exon 25.
Runkel F, Büssow H, Seburn KL, Cox GA, Ward DM, Kaplan J, Franz T. Runkel F, et al. Mamm Genome. 2006 Mar;17(3):203-10. doi: 10.1007/s00335-005-0015-1. Epub 2006 Mar 3. Mamm Genome. 2006. PMID: 16518687 - Identification of mutations in two major mRNA isoforms of the Chediak-Higashi syndrome gene in human and mouse.
Barbosa MD, Barrat FJ, Tchernev VT, Nguyen QA, Mishra VS, Colman SD, Pastural E, Dufourcq-Lagelouse R, Fischer A, Holcombe RF, Wallace MR, Brandt SJ, de Saint Basile G, Kingsmore SF. Barbosa MD, et al. Hum Mol Genet. 1997 Jul;6(7):1091-8. doi: 10.1093/hmg/6.7.1091. Hum Mol Genet. 1997. PMID: 9215680 Free PMC article. - Cloning of bovine LYST gene and identification of a missense mutation associated with Chediak-Higashi syndrome of cattle.
Kunieda T, Nakagiri M, Takami M, Ide H, Ogawa H. Kunieda T, et al. Mamm Genome. 1999 Dec;10(12):1146-9. doi: 10.1007/s003359901181. Mamm Genome. 1999. PMID: 10594238 - Spectrum of LYST mutations in Chediak-Higashi syndrome: a report of novel variants and a comprehensive review of the literature.
Morimoto M, Nicoli ER, Kuptanon C, Roney JC, Serra-Vinardell J, Sharma P, Adams DR, Gallin JI, Holland SM, Rosenzweig SD, Barbot J, Ciccone C, Huizing M, Toro C, Gahl WA, Introne WJ, Malicdan MCV. Morimoto M, et al. J Med Genet. 2024 Feb 21;61(3):212-223. doi: 10.1136/jmg-2023-109420. J Med Genet. 2024. PMID: 37788905 Review. - Clinical, molecular, and cell biological aspects of Chediak-Higashi syndrome.
Introne W, Boissy RE, Gahl WA. Introne W, et al. Mol Genet Metab. 1999 Oct;68(2):283-303. doi: 10.1006/mgme.1999.2927. Mol Genet Metab. 1999. PMID: 10527680 Review.
Cited by
- Lymphocytes with cytotoxic activity induce rapid microtubule axonal destabilization independently and before signs of neuronal death.
Miller NM, Shriver LP, Bodiga VL, Ray A, Basu S, Ahuja R, Jana A, Pahan K, Dittel BN. Miller NM, et al. ASN Neuro. 2013 Feb 6;5(1):e00105. doi: 10.1042/AN20120087. ASN Neuro. 2013. PMID: 23289514 Free PMC article. - Efficacy of T-cell assays for the diagnosis of primary defects in cytotoxic lymphocyte exocytosis.
Chiang SCC, Covill LE, Tesi B, Campbell TM, Schlums H, Nejati-Zendegani J, Mördrup K, Wood S, Theorell J, Sekine T, Al-Herz W, Akar HH, Belen FB, Chan MY, Devecioglu O, Aksu T, Ifversen M, Malinowska I, Sabel M, Unal E, Unal S, Introne WJ, Krzewski K, Gilmour KC, Ehl S, Ljunggren HG, Nordenskjöld M, Horne A, Henter JI, Meeths M, Bryceson YT. Chiang SCC, et al. Blood. 2024 Aug 22;144(8):873-887. doi: 10.1182/blood.2024024499. Blood. 2024. PMID: 38958468 Free PMC article. - Susceptibility of lysosomes to rupture is a determinant for plasma membrane disruption in tumor necrosis factor alpha-induced cell death.
Ono K, Kim SO, Han J. Ono K, et al. Mol Cell Biol. 2003 Jan;23(2):665-76. doi: 10.1128/MCB.23.2.665-676.2003. Mol Cell Biol. 2003. PMID: 12509464 Free PMC article. - Clinical, laboratory and molecular signs of immunodeficiency in patients with partial oculo-cutaneous albinism.
Dotta L, Parolini S, Prandini A, Tabellini G, Antolini M, Kingsmore SF, Badolato R. Dotta L, et al. Orphanet J Rare Dis. 2013 Oct 17;8:168. doi: 10.1186/1750-1172-8-168. Orphanet J Rare Dis. 2013. PMID: 24134793 Free PMC article. Review. - Insights into NK cell biology from human genetics and disease associations.
Wood SM, Ljunggren HG, Bryceson YT. Wood SM, et al. Cell Mol Life Sci. 2011 Nov;68(21):3479-93. doi: 10.1007/s00018-011-0799-y. Epub 2011 Aug 27. Cell Mol Life Sci. 2011. PMID: 21874350 Free PMC article. Review.
References
- Blume RS, Wolff SM. Med. Baltimore. 1972;51:247–280. - PubMed
- Zhao H, et al. Lab. Invest. 1994;71:25–34. - PubMed
- Willingham MC, Spicer SS, Vincent RA. Expl Cell Res. 1981;136:157–168. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous