Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism - PubMed (original) (raw)
Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism
Z Xia et al. J Neurosci. 1996.
Abstract
The regulation of gene expression by neurotransmitters is likely to play a key role in neuroplasticity both during development and in the adult animal. Therefore, it is important to determine the mechanisms of neuronal gene regulation to understand fully the mechanisms of learning, memory, and other long-term adaptive changes in neurons. The neurotransmitter glutamate stimulates rapid and transient induction of many genes, including the c-fos proto-oncogene. The c-fos promoter contains several critical regulatory elements, including the serum response element (SRE), that mediate glutamate-induced transcription in neurons; however, the mechanism by which the SRE functions in neurons has not been defined. In this study, we sought to identify transcription factors that mediate glutamate induction of transcription through the SRE in cortical neurons and to elucidate the mechanism(s) of transcriptional activation by these factors. To facilitate this analysis, we developed an improved calcium phosphate coprecipitation procedure to transiently introduce DNA into primary neurons, both efficiently and consistently. Using this protocol, we demonstrate that the transcription factors serum response factor (SRF) and Elk-1 can mediate glutamate induction of transcription through the SRE in cortical neurons. There are at least two distinct pathways by which glutamate signals through the SRE: an SRF-dependent pathway that can operate in the absence of Elk and an Elk-dependent pathway. Activation of the Elk-dependent pathway of transcription seems to require phosphorylation of Elk-1 by extracellular signal-regulated kinases (ERKs), providing evidence for a physiological function of ERKs in glutamate signaling in neurons. Taken together, these findings suggest that SRF, Elk, and ERKs may have important roles in neuroplasticity.
Figures
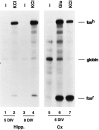
Fig. 2.
Genes transfected into neurons are correctly regulated in response to extracellular stimuli. The plasmid pF4, which contains the intact human c-fos gene including 750 bp of 5′ regulatory sequence, was transfected 3 d after plating (3 DIV) into hippocampal (Hipp.,lanes 1–4) or cortical neurons (Cx,lanes 5–7). The plasmid pSVα1, which encodes the human α-globin gene under the control of the SV40 promoter, was cotransfected as an internal control for transfection efficiency and RNA recovery (2 μg/plate). For hippocampal neurons, 0.9 × 106 cells were plated onto each 60-mm-diameter plate, and 5 μg of pF4 plasmid DNA was used for transfection. For cortical neurons, 3 × 106 cells were plated onto each 60-mm-diameter plate, and 4 μg of pF4 plasmid DNA was used for transfection. Transfected cells were either left untreated (−) or stimulated with 55 m
m
KCl or 10 μ
m
glutamate (Glu) on 5 DIV (_lanes 1_and 2), 6 DIV (lanes 5–7), or 8 DIV (_lanes 3_and 4). RNase protection analysis was used to measure the expression of the transfected human c-fos gene (fosh), the human α-globin gene, and the endogenous rat c-fos gene (fosr).
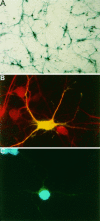
Fig. 1.
Transient transfection of the β-galactosidase gene into primary cortical neurons using a modified calcium phosphate procedure. A, X-Gal staining (blue) of cortical neurons (P0) transfected at 3 DIV with an expression vector encoding β-galactosidase. B, A representative immunofluorescence photomicrograph of a neuron transfected with β-galactosidase. Transfected cortical neurons cultured from E17/18 rats were detected by immunostaining with a monoclonal antibody to β-galactosidase, visualized by fluorescein-conjugated goat antibody to mouse IgG (green). The cells were coimmunostained for the neuronal marker protein MAP-2, which was visualized using a Texas Red-conjugated goat antibody to rabbit IgG (red). The transfected neuron appears yellow because of the colocalization of green staining from anti-β-galactosidase and red staining from anti-MAP-2.C, A representative immunofluorescence photomicrograph of a healthy neuron expressing β-galactosidase. A transfected E17/18 rat cortical neuron was detected as in B. The nucleus was visualized using the DNA dye Hoechst 33258 (blue). The evenly stained, round morphology is typical of a healthy (nonapoptotic) nucleus.
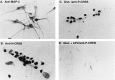
Fig. 3.
Glutamate responsiveness of cultured cortical neurons. Cortical neurons were immunostained with an antibody to the neuronal marker protein MAP-2 (A), an antibody to the N terminus of CREB (B), or an antibody specific for CREB that is phosphorylated at serine133 (C, D). Cells were unstimulated (A, B), or stimulated for 10 min with 10 μ
m
glutamate in the absence (C) or presence (D) of 100 μ
m
APV. Glutamate induced CREB phosphorylation only in cells with neuronal morphology.
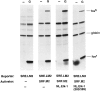
Fig. 4.
Contribution of the SRF and TCF binding sites for glutamate stimulation of SRE-mediated transcription. A, Glutamate stimulation of SRE-mediated transcription is dependent on the activation of NMDA receptors. Cortical neurons (3 × 106 cells/plate) were transfected with the wild-type c-fos SRE-containing construct pAF42. SRE.wt (1 μg/plate) at 3 DIV. Three days later, cells were left untreated (−), treated with 10 μ
m
glutamate in the absence or presence of 100 μ
m
APV, or treated with 100 μ
m
NMDA. RNase protection analysis was used to measure the expression of the transfected human c-fos gene (fosh), the human α-globin gene, and the endogenous rat c-fos gene (fosr). B, Effects of mutations within the SRE that interfere with the binding of SRF or TCFs on SRE-mediated transcription. Cortical neurons (3 × 106 cells/plate) were transfected with 1 μg of pAF42.SRE.wt, or constructs with mutations in the SRE that disrupt SRF binding (pAF42.SRE.pm1 or
pAF42.SRE.mut2
), or TCF binding (
pAF42.SRE.mut6
). Three days later, the transfected cells were either left untreated (−) or stimulated with 10 μ
m
glutamate. Similar results were obtained from three independent experiments. The relative levels of transcription between different reporter constructs after glutamate treatment were determined by the ratio of fosh/globin.
Fig. 5.
Ectopically expressed SRF can mediate SRE-dependent transcription stimulated by glutamate. A, Expression vectors used in the altered binding specificity assay (Hill et al., 1993). Amino acids 133–166 of the DNA binding domain of SRF were replaced with 33 amino acids from the DNA binding domain of MCM1 (black box) to create SRF.M2. Amino acids 3–87 of the LexA DNA binding domain (dotted box) were fused in-frame in front of amino acid 107 of Elk-1, which eliminates the Elk-1 DNA binding domain, to generate LexA-Elk. Ten additional amino acids encoding a nuclear localization signal (crossed box) were added to the N terminus of LexA-Elk to ensure its nuclear localization (NL.Elk-1). B, Reporter constructs used in the altered binding specificity assay (Hill et al., 1993; Miranti et al., 1995). Endogenous SRF binds as a homodimer to the CArG box of the wild-type SRE (pAF42.SRE.wt). TCFs, e.g., Elk-1, bind as a monomer to an ETS sequence adjacent to the CArG box only when SRF is already bound to the SRE. The CArG site was replaced by an MCM1 promoter sequence to generate pAF42.SRE.M2, which can bind SRF.M2 but not wild-type endogenous SRF. Endogenous Elk-like proteins can still bind to the ETS site in the SRE.M2. Additional substitution of the ETS site with a LexA site generated pAF42.SRE.LM2, which contains mutations in both the SRF and Elk binding sites and is therefore incapable of binding endogenous SRF or Elk. Elk binding activity can be restored to the SRE.LM2 promoter element by NL.Elk-1 when SRF.M2 is present. Reporters with two adjacent copies of the mutated SREs were used. C, SRF can mediate SRE-dependent transcription in response to glutamate. Cortical neurons were transfected with the reporter construct pAF42.SRE.M2 either alone or together with an expression vector encoding SRF.M2. Three days later, cells were either left untreated (−) or treated with 10 μ
m
glutamate. The level of mRNA transcribed from the reporter plasmid was assessed as described in the legend to Figure 4. Similar results were obtained from three independent experiments.
Fig. 6.
Glutamate stimulates SRE-mediated transcription through an Elk-dependent pathway. Cortical neurons (3 × 106 cells/plate) were transfected with the c-fos reporter pAF42.SRE.LM2 (1 μg) together with 200 ng of various expression plasmids as indicated: SRF.M2, NL.Elk-1, or NL.Elk-1(383/389). NL.Elk-1(383/389) contains alanine substitutions at Ser383 and Ser389, two of the MAP kinase phosphorylation sites in the C terminus of Elk-1. Three days after transfection, cells were either left untreated (−) or treated with 10 μ
m
glutamate. The level of mRNA transcribed from the reporter plasmid was assessed and quantitated as described in the legend for Figure 4. Relative levels of transcription from the reporter pAF42.SRE.LM2 after glutamate stimulation are SRF.M2, 100%; SRF.M2 + NL.Elk-1, 154 ± 5% (SEM,n = 5); SRF.M2 + NL.Elk-1(383/389), 104 ± 12% (SEM, n = 3).
Fig. 7.
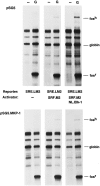
Coexpression of a MAP kinase phosphatase MKP-1 blocks Elk-dependent transcription. Cortical neurons were transfected with the c-fos reporter pAF42.SRE.LM2 alone or together with expression plasmids encoding SRF.M2 or NL.Elk-1, as indicated. Cells were also cotransfected with either the empty cloning vector pSG5 (top) or the expression vector pSG5.MKP-1 (bottom). Three days later, cells were either left untreated (−) or treated with 10 μ
m
glutamate. The level of mRNA transcribed from the reporter plasmid was determined as described in the legend to Figure 4. In cells expressing MKP-1 (bottom), coexpression of Elk-1 failed to enhance transcription over that obtained with SRF alone, in comparison to cells transfected with vector alone (top). Similar results were obtained from two independent experiments.
Similar articles
- Calcium controls gene expression via three distinct pathways that can function independently of the Ras/mitogen-activated protein kinases (ERKs) signaling cascade.
Johnson CM, Hill CS, Chawla S, Treisman R, Bading H. Johnson CM, et al. J Neurosci. 1997 Aug 15;17(16):6189-202. doi: 10.1523/JNEUROSCI.17-16-06189.1997. J Neurosci. 1997. PMID: 9236230 Free PMC article. - Glutamate induces phosphorylation of Elk-1 and CREB, along with c-fos activation, via an extracellular signal-regulated kinase-dependent pathway in brain slices.
Vanhoutte P, Barnier JV, Guibert B, Pagès C, Besson MJ, Hipskind RA, Caboche J. Vanhoutte P, et al. Mol Cell Biol. 1999 Jan;19(1):136-46. doi: 10.1128/MCB.19.1.136. Mol Cell Biol. 1999. PMID: 9858538 Free PMC article. - Growth hormone stimulates phosphorylation and activation of elk-1 and expression of c-fos, egr-1, and junB through activation of extracellular signal-regulated kinases 1 and 2.
Hodge C, Liao J, Stofega M, Guan K, Carter-Su C, Schwartz J. Hodge C, et al. J Biol Chem. 1998 Nov 20;273(47):31327-36. doi: 10.1074/jbc.273.47.31327. J Biol Chem. 1998. PMID: 9813041 - The role of regulated phosphorylation in the biological activity of transcription factors SRF and Elk-1/SAP-1.
Papavassiliou AG. Papavassiliou AG. Anticancer Res. 1994 Sep-Oct;14(5A):1923-6. Anticancer Res. 1994. PMID: 7847828 Review. - Critical Protein-Protein Interactions Determine the Biological Activity of Elk-1, a Master Regulator of Stimulus-Induced Gene Transcription.
Thiel G, Backes TM, Guethlein LA, Rössler OG. Thiel G, et al. Molecules. 2021 Oct 11;26(20):6125. doi: 10.3390/molecules26206125. Molecules. 2021. PMID: 34684708 Free PMC article. Review.
Cited by
- Improved detection of electrical activity with a voltage probe based on a voltage-sensing phosphatase.
Tsutsui H, Jinno Y, Tomita A, Niino Y, Yamada Y, Mikoshiba K, Miyawaki A, Okamura Y. Tsutsui H, et al. J Physiol. 2013 Sep 15;591(18):4427-37. doi: 10.1113/jphysiol.2013.257048. Epub 2013 Jul 8. J Physiol. 2013. PMID: 23836686 Free PMC article. - The Three Musketeers in the Medial Prefrontal Cortex: Subregion-specific Structural and Functional Plasticity Underlying Fear Memory Stages.
Sung Y, Kaang BK. Sung Y, et al. Exp Neurobiol. 2022 Aug 31;31(4):221-231. doi: 10.5607/en22012. Exp Neurobiol. 2022. PMID: 36050222 Free PMC article. Review. - Arsenite-induced apoptosis in cortical neurons is mediated by c-Jun N-terminal protein kinase 3 and p38 mitogen-activated protein kinase.
Namgung U, Xia Z. Namgung U, et al. J Neurosci. 2000 Sep 1;20(17):6442-51. doi: 10.1523/JNEUROSCI.20-17-06442.2000. J Neurosci. 2000. PMID: 10964950 Free PMC article. - P2X receptor trafficking in neurons is subunit specific.
Bobanovic LK, Royle SJ, Murrell-Lagnado RD. Bobanovic LK, et al. J Neurosci. 2002 Jun 15;22(12):4814-24. doi: 10.1523/JNEUROSCI.22-12-04814.2002. J Neurosci. 2002. PMID: 12077178 Free PMC article. - Upstream stimulatory factors are mediators of Ca2+-responsive transcription in neurons.
Chen WG, West AE, Tao X, Corfas G, Szentirmay MN, Sawadogo M, Vinson C, Greenberg ME. Chen WG, et al. J Neurosci. 2003 Apr 1;23(7):2572-81. doi: 10.1523/JNEUROSCI.23-07-02572.2003. J Neurosci. 2003. PMID: 12684442 Free PMC article.
References
- Alessi DR, Smythe C, Keyse SM. The human CL100 gene encodes a Tyr/Thr-protein phosphatase which potently and specifically inactivates MAP kinase and suppresses its activation by oncogenic ras in Xenopus oocyte extracts. Oncogene. 1993;8:2015–2020. - PubMed
- Bading H, Greenberg ME. Stimulation of protein tyrosine phosphorylation by NMDA receptor activation. Science. 1991;253:912–914. - PubMed
- Bading H, Ginty DD, Greenberg ME. Regulation of gene expression in hippocampal neurons by distinct calcium signaling pathways. Science. 1993;260:181–186. - PubMed
- Bean BP. Classes of calcium channels in vertebrate cells. Annu Rev Physiol. 1989;51:367–384. - PubMed
- Bear MF, Malenka RC. Synaptic plasticity: LTP and LTD. Curr Opin Neurobiol. 1994;4:389–399. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 NS028829/NS/NINDS NIH HHS/United States
- NS 07009-2021/NS/NINDS NIH HHS/United States
- NS28829/NS/NINDS NIH HHS/United States
- R01 NS028829/NS/NINDS NIH HHS/United States
- P30 HD018655/HD/NICHD NIH HHS/United States
- P30-HD 18655/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous