A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3' end - PubMed (original) (raw)
A novel RNA polymerase I-dependent RNase activity that shortens nascent transcripts from the 3' end
H Tschochner. Proc Natl Acad Sci U S A. 1996.
Abstract
A novel RNase activity was identified in a yeast RNA polymerase I (pol I) in vitro transcription system. Transcript cleavage occurred at the 3' end and was dependent on the presence of ternary pol I/DNA/RNA complexes and an additional protein factor not identical to transcription factor IIS (TFIIS). Transcript cleavage was observed both on arrested complexes at the linearized ends of the transcribed DNA and on intrinsic blocks of the DNA template. Shortened transcripts that remained associated within the ternary complexes were capable of resuming RNA chain elongation. Possible functions of the nuclease for transcript elongation or termination are discussed.
Figures
Figure 1
Transcripts are cleaved from the 3′ end. (A) Primer extension. Pol I-dependent transcripts were generated in the absence (lane 1) or presence (lane 2) of fraction 41 (Resource S column) and reverse-transcribed using a32P-labeled oligonucleotide (on 5-115) as the primer. A DNA sequencing reaction with the same oligonucleotide as the primer and pSES5 as the template was performed independently. The resulting DNA fragments were separated on a 7% polyacrylamide sequencing gel. The correct start site of pol I-dependent transcription is indicated by an arrow. (The first 8 nt are 5′-AUGCGAAA.) (B) S-1 nuclease mapping of the 3′ end. Transcription from pSES5 linearized with the enzymes indicated was performed in the presence of [32P]GTP. Isolated radiolabeled RNA was hybridized to oligonucleotide S-13p and subjected to S-1 nuclease treatment. Protected fragments were visualized by autoradiography of 13% polyacrylamide/7 M urea gels. As schematically detailed, RNA transcribed from the _Bam_HI-linearized pSES5 and hybridized to the oligonucleotide should lead to a completely protected fragment 64 nt long (lanes 3 and 4, respectively, and 3/4 in the scheme). In contrast, a shorter fragment should result from the hybridization of the same oligonucleotide with RNA transcribed from the_Eco_RV-linearized template (lane 1 and 1 in the scheme). If RNA cleavage takes place at the 3′ end of the transcript, the addition of fraction should lead to an even shorter protected fragment (lane 2 and 2 in the respective representation). Positions of marker oligonucleotides are indicated at the right. (C) Analysis of run-off transcripts on a high-resolution gel. Accurate initiated transcripts were synthesized in vitro in the absence (lane 1) or presence (lane 2) of fraction 41 and separated on a 7% denaturing polyacrylamide sequencing gel. A purin/pyrimidine ladder is shown on the right. (D) Time course of transcript cleavage. After the generation of full-length transcripts by the pol I complex for 20 min, fraction 41 was added. Equal amounts of the reaction were removed and stopped at time points indicated.
Figure 2
RNA cleavage is not sequence-specific. Transcription assays with different run-off templates. Plasmids pSES5 and pSKpI were linearized with different restriction enzymes as indicated. The resulting DNA served as templates for promoter-specific pol I-dependent transcription in the absence or presence of fraction 41. The resulting transcripts were separated on a 7% polyacrylamide sequencing gel. Note that _Eco_RV, _Bam_HI, and_Sac_I produce blunted, 5′, and 3′ overhung ends on the templates.
Figure 3
A ternary complex is essential to enable RNA cleavage in pol I-dependent transcription assays. (A Upper) Transcript cleavage reaction is restricted to ternary complexes. Run-off transcription was performed with immobilized template for 20 min. After separating the supernatant from the pellet, fraction 41 was added to both fractions (lanes 3 and 4). In control reactions fraction 41 was added (lane 2) or omitted (lane 1) to transcription assays with completed transcripts generated from DNA template in solution. The position of uncleaved transcripts is indicated (tr pol I). (A Lower) 32P-labeled run-off transcripts generated from the pSES5/_Bam_HI fragment were isolated after DNase and proteinase K treatment. Aliquots were added to transcription reactions containing pSES5/_Eco_RV as the template in the presence of 1.5 μl (lane 3) and 3 μl (lane 4) of fraction 41. Control reactions show transcripts derived from the pSES5/_Bam_HI and pSES5/_Eco_RV template when processed in the absence (lane 1) or presence (lane 2) of fraction 41. Positions of the different uncleaved transcripts are indicated (tr _Bam_HI and tr_Eco_RV). (B Upper) Cleavage factor distinguishes between two different cotranscriptionally synthesized RNA. Increasing amounts of fraction 41 (lanes 2–4) were added to specific initiated run-off transcription assays in which mitochondrial RNA polymerase and pol I simultanously transcribed the same template. Positions of the corresponding transcripts are indicated (tr mito, accurately initiated transcript synthesized by mitochondrial polymerase; tr pol I, accurately initiated pol I-specific transcript). Although equal amounts of nonspecific RNA-synthesizing activities were used (data not shown), transcripts due to mitochondrial polymerase predominated, probably because the Mg2+ and potassium acetate concentration was suboptimal for pol I-dependent transcription. (B Lower) Transcripts generated by mitochondrial RNA polymerase are associated with the template and cannot be cleaved by fraction 41. Run-off transcription with the immobilized template was performed by using the mitochondrial RNA polymerase complex for 20 min. After separating the supernatant (s) and pellet (p), fraction 41 was added (lanes 3 and 4) or omitted (lanes 1 and 2).
Figure 4
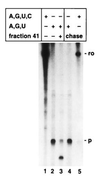
Cleaved transcripts that remain associated within ternary complexes can be reextended upon addition of NTP. Transcription reactions on 3′-extended immobilized templates (pItailKS) were performed as described. In the presence of all four NTPs a 188-nt-long run-off RNA (ro) was generated (lane 1). In the absence of CTP (C), transcription paused at the first CTP to be incorporated (p) and revealed a RNA fragment 53 nt long (lane 2). Purified and washed ternary complexes were incubated without (lane 2) or with (lanes 3–5) fraction 41 for 20 min in the absence of NTPs. After washing the immobilized templates and associated complexes, the complexes were allowed to resume elongation for 20 min at 25°C providing ATP (A), GTP (G), and UTP (U) (lane 4) or all four NTPs (lane 5). RNA fragments were separated on a 7.5% acrylamide/7 M urea gel.
Figure 5
Cleavage reaction is not dependent on TFIIS. (Top) In vitro transcription assays without (lane 1) or with (lane 2) fraction 41 and with increasing amounts of purified recombinant yTFIIS. Recombinant TFIIS was more than 95% pure (generous gift from A. Edwards, Hamilton, Canada). Recombinant TFIIS (5 ng) was able to stimulate pol II-dependent RNA synthesis (data not shown). (Middle) In vitro transcription assays in the presence of fraction 41, which has been immunodepleted by anti-TFIIS antibodies (lanes 4–6) or the corresponding preimmunserum (pre) (lanes 1–3). (Bottom) Immunoblot with antibodies against yTFIIS. Equal amounts of fraction 41 were immunodepleted by an anti-TFIIS antiserum (lanes 4 and 5) and the corresponding preimmunantiserum (lanes 2 and 3). The indicated volumes were subjected to electrophoresis in a 12% SDS/polyacrylamide gel, electroblotted onto nitrocellulose, and probed with anti-TFIIS antibodies. Lane 1 shows 10 ng recombinant purified yTFIIS.
Similar articles
- The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination.
Chédin S, Riva M, Schultz P, Sentenac A, Carles C. Chédin S, et al. Genes Dev. 1998 Dec 15;12(24):3857-71. doi: 10.1101/gad.12.24.3857. Genes Dev. 1998. PMID: 9869639 Free PMC article. - Purified yeast RNA polymerase II reads through intrinsic blocks to elongation in response to the yeast TFIIS analogue, P37.
Christie KR, Awrey DE, Edwards AM, Kane CM. Christie KR, et al. J Biol Chem. 1994 Jan 14;269(2):936-43. J Biol Chem. 1994. PMID: 8288647 - TFIIS binds to mouse RNA polymerase I and stimulates transcript elongation and hydrolytic cleavage of nascent rRNA.
Schnapp G, Graveley BR, Grummt I. Schnapp G, et al. Mol Gen Genet. 1996 Sep 25;252(4):412-9. doi: 10.1007/BF02173006. Mol Gen Genet. 1996. PMID: 8879242 - The mechanism of transcription termination by RNA polymerase I.
Reeder RH, Lang W. Reeder RH, et al. Mol Microbiol. 1994 Apr;12(1):11-5. doi: 10.1111/j.1365-2958.1994.tb00989.x. Mol Microbiol. 1994. PMID: 8057832 Review. - Promoting elongation with transcript cleavage stimulatory factors.
Fish RN, Kane CM. Fish RN, et al. Biochim Biophys Acta. 2002 Sep 13;1577(2):287-307. doi: 10.1016/s0167-4781(02)00459-1. Biochim Biophys Acta. 2002. PMID: 12213659 Review.
Cited by
- La protein and its associated small nuclear and nucleolar precursor RNAs.
Maraia RJ, Intine RV. Maraia RJ, et al. Gene Expr. 2002;10(1-2):41-57. Gene Expr. 2002. PMID: 11868987 Free PMC article. Review. - Multisubunit DNA-Dependent RNA Polymerases from Vaccinia Virus and Other Nucleocytoplasmic Large-DNA Viruses: Impressions from the Age of Structure.
Mirzakhanyan Y, Gershon PD. Mirzakhanyan Y, et al. Microbiol Mol Biol Rev. 2017 Jul 12;81(3):e00010-17. doi: 10.1128/MMBR.00010-17. Print 2017 Sep. Microbiol Mol Biol Rev. 2017. PMID: 28701329 Free PMC article. Review. - Selectivity and proofreading both contribute significantly to the fidelity of RNA polymerase III transcription.
Alic N, Ayoub N, Landrieux E, Favry E, Baudouin-Cornu P, Riva M, Carles C. Alic N, et al. Proc Natl Acad Sci U S A. 2007 Jun 19;104(25):10400-5. doi: 10.1073/pnas.0704116104. Epub 2007 Jun 6. Proc Natl Acad Sci U S A. 2007. PMID: 17553959 Free PMC article. - RNA polymerase I structure and transcription regulation.
Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. Engel C, et al. Nature. 2013 Oct 31;502(7473):650-5. doi: 10.1038/nature12712. Epub 2013 Oct 23. Nature. 2013. PMID: 24153182 - Genetic analyses led to the discovery of a super-active mutant of the RNA polymerase I.
Darrière T, Pilsl M, Sarthou MK, Chauvier A, Genty T, Audibert S, Dez C, Léger-Silvestre I, Normand C, Henras AK, Kwapisz M, Calvo O, Fernández-Tornero C, Tschochner H, Gadal O. Darrière T, et al. PLoS Genet. 2019 May 28;15(5):e1008157. doi: 10.1371/journal.pgen.1008157. eCollection 2019 May. PLoS Genet. 2019. PMID: 31136569 Free PMC article.
References
- Borukhov S, Sagitov V, Goldfarb A. Cell. 1993;72:459–466. - PubMed
- Nudler E, Goldfarb A, Kashlev M. Science. 1994;265:793–796. - PubMed
- Arndt K M, Chamberlin M J. J Mol Biol. 1990;213:79–108. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases